Chapter 5
Running makemap Outside the Pipeline
The previous chapter described how to make maps using the SCUBA-2 Pipeline. Users—particularly
new users—are encouraged to use the pipeline for their map-making. However, greater control of
the map-making process is available by running the makemap command directly, rather than
from within the pipeline. This chapter describes how to do this, but should be seen as “advanced
usage”.
Note, when makemap is run manually (rather than from the pipeline) the atmospheric extinction
correction is applied automatically (see Appendix B), but the resulting image will be in units of pW,
not Jy. Each pixel value in the map represents the weighted mean value of the bolometer samples (in
pW) that fall within the pixel, removal of the noise components described in Section 3.3. Each weight
is the reciprocal of the variance associated with the bolometer. Thus each pixel value can be thought
of as the astronomical power received by a typical bolometer at each point on the sky. Section 8.1
describes how to convert a map from units of pW to Jy; see Appendices B, E, and F for more
information.
5.1 Running makemap
Before running makemap directly, you need to ensure that the Starlink environment has been initialised and
the Smurf package started (see Section 1.3.2 and Section 1.3.3).
To run makemap, you need to supply values for the following command-line
parameters:
IN | A list of input NDFs containing raw SCUBA-2 data (see Section 2.3 and Section 1.3.1). There are
many ways in which the list of files can be supplied, as described in Section “Specifying Groups of
Objects” in SUN/95. The easiest is to create a simple text file containing the names of the raw data
files – one per line—and then supply the name of the text file, preceded by an up-caret character
( ^ ), as the value for parameter IN. Note, the names of the raw data files can contain wild-cards
such as “”
and “?”. See below for examples.
|
|---|
OUT | The name of the NDF in which to store the final map. The supplied file name should either have
a file type of “.sdf”, or no file type at all (in which case .sdf will be appended to the supplied
value). Any existing file with the same name will be over-written.
|
|---|
CONFIG | A string that specifies new values for one or more configuration parameters. These new values
are used in place of the default values described in SUN/258. Any configuration parameter not
specified by the CONFIG string retains its default value. As with files names, there are many ways
in which the group of parameter values can be specified, and the same documentation should be
consulted for details (SUN/95). Again, the easiest way is to list the parameter values in a simple
text file with one parameter setting (a “<name>=<value>” string) on each line. See Section 3.7 for
a list of the pre-defined configuration files that come with Smurf. Note, CONFIG can be set to the
special value “def” to force makemap to use the default values for all parameters. It is possible to
create a map using just these default values, but it will not usually be a very good map. Advice on
which parameters to edit can be found in Section 6.
|
|---|
If any of the above parameters are not given on the makemap command line, prompts will be issued and you will
be invited to supply values for the missing parameters.
So an example command line would be:
% makemap in=^myfiles.lis out=850_crl2688 \
config=^$STARLINK_DIR/share/smurf/dimmconfig_bright_compact.lis
Here, the file myfiles.lis contains a list of the raw data files to be included in the map, and could for instance
look like this:
% cat myfiles.lis
/jcmtdata/raw/scuba2/s8a/20120720/00030/*
/jcmtdata/raw/scuba2/s8b/20120720/00030/*
/jcmtdata/raw/scuba2/s8c/20120720/00030/*
/jcmtdata/raw/scuba2/s8d/20120720/00030/*
This uses all available data for all four 850 m
sub-arrays, for observation 30 taken on 20th July
2012.
The file dimmconfig_bright_compact is one of the pre-defined configuration files supplied with Smurf. It
contains configuration parameter values that are optimised for creating maps from bright compact
sources.
Tip: An up-caret (
^ ) is required any time you are reading in a group text file in
Starlink. For the map-maker this
includes the configuration file (a group of configuration parameters) and the list of input files ( a group of
NDFs—
e.g. in=^myfiles.lis).
After the makemap command completes, the final map will be left in file 850_crl2688.sdf and can be displayed
using Gaia:
There are many other command-line parameters that can be supplied when running makemap, but only the three
listed above are mandatory. All the others assume default values if not supplied on the command-line. For a full
list of all available command-line parameters, their functions and default values, see the description of
makemap within the smurf document, SUN/258.
You can supply values for any of these extra parameters on the command line. For instance, one of the
command-line parameters that is usually defaulted is PIXSIZE, which specifies the pixel size for the final map,
in arcseconds. The default pixel sizes, defined as one quarter of the Airy disk rounded up to the nearest half
arcsecond, are:
- 2 arcsec at 450m
- 4 arcsec at 850m
So for instance, the above command-line could be modified as follows to produce a map with 3-arcsecond
pixels:
% makemap in=^myfiles.lis out=850_crl2688 pixsize=3 \
config=^$STARLINK_DIR/share/smurf_bright_compact.lis
Note, if parameters are specified by name on the command-line, as in these examples, then the order in which they are specified
is insignificant.
Also, parameter names are case-insensitive.
Tip: Map-maker not finding your raw data files from a path? Check you have escaped or protected any shell
meta-characters in your ‘in’ value, for instance by putting double quotes around it or escaping wild-cards
using a backslash (e.g. “in=\*.sdf” or just “in=\*”). Note, this issue applies only to wild-cards
included directly on the command line—there is no need to escape or protect wild cards within a text
file.
5.2 Interpreting the screen output from makemap
Whilst makemap runs, it outputs information continuously to the screen about what it is doing. The amount of
information displayed can be controlled using the MSG_FILTER command-line parameter. It defaults to
“normal”, but if you want more information you could set it to (say) “verbose”:
% makemap in=^myfiles.lis out=850_crl2688 msg_filter=verb \
config=^$STARLINK_DIR/share/smurf_bright_compact.lis
Note—unambiguous abbreviations may be used for many command line parameters—so “verb” is acceptable
instead of “verbose”. Setting MSG_FILTER to “quiet” will suppress all screen output.
Tip: Map-maker generates a lot of screen output! The main things to check are that the “
NORMALIZED
MAP CHANGE” value (the mean value, not the max value) decreases nicely towards your requested
maptol value as each iteration is completed, and that the “
Total samples available for map:”
value does not fall too low (you should usually be looking for values above 50% of maximum). If
neither of these two items look problematic, it is usually safe to pay less attention to the other screen
output.
The following shows the screen output generated by a typical run of makemap if MSG_FILTER is left set to its
default value of “normal”. Explanatory comments, which are not actually part of the output generated by
makemap, are included in emphasised type. The input data are a short observation of a bright compact source
(CRL 2688):
% makemap in=^myfiles.lis out=850_crl2688 \
config=^$STARLINK_DIR/share/smurf_bright_compact.lis
Out of 32 input files, 4 were darks, 8 were fast flats and 20 were science
Processing data from instrument ’SCUBA-2’ for object ’CRL2688’ from the
following observation :
20120720 #30 scan
The output starts by reporting information about the input data files, including the number that contain on-sky bolometer
values (“science” data), the astronomical object and the SCUBA-2 observation number.
MAKEMAP: Map-maker will use no more than 92586 MiB of memory
Projection parameters used:
CRPIX1 = 0
CRPIX2 = 0
CRVAL1 = 315.578333333333 ( RA = 21:02:18.800 )
CRVAL2 = 36.6938055555556 ( Dec = 36:41:37.70 )
CDELT1 = -0.00111111111111111 ( -4 arcsec )
CDELT2 = 0.00111111111111111 ( 4 arcsec )
CROTA2 = 0
Output map pixel bounds: ( -132:122, -126:129 )
Output map WCS bounds:
Right ascension: 21:01:38.318 -> 21:03:03.280
Declination: 36:33:07.19 -> 36:50:11.70
Next comes information about the output map. The world coordinate system is described by means of the equivalent FITS-WCS
keywords
The reported pixel bounds of the output map refer to a pixel coordinate system in which the source is centred at position
(0,0). Note, the definition of pixel coordinates within the NDF format allows the origin of pixel coordinates to be at any
nominated position within the array—it does not have to be at the bottom left corner as in FITS—and makemap chooses to
put the pixel origin at the specified source position.
Finally, the bounds of the map are given in the celestial coordinate system specified by the SYSTEM command-line
parameter. This parameter defaults to “tracking”, which causes the map to be created in the celestial coordinate system in
which the observation parameters were originally defined. It may instead be set to a specific coordinate system (e.g.
“galactic”, “icrs”, etc.) to force the map to be made in that system.
smf_iteratemap: will down-sample data to match angular scale of 4 arcsec
smf_iteratemap: Iterate to convergence (max 40)
smf_iteratemap: stop when mean normalized map change < 0.05
By default, the data are down-sampled so that the on-sky distance between adjacent samples is roughly equal to the
pixel size (4 arcseconds in this case). This saves memory and computing time without adversely affecting
the final map. The degree of down-sampling can be controlled using the downsampscale configuration
parameter.
Next comes information about the stopping criteria for the iterative map-making algorithm. In this case,
iterations will stop when 40 iterations are completed, or the normalised change in the map between iterations
reduces to less that 0.05. These values are specified by configuration parameters numiter and maptol (see
Section 3.4).
smf_iteratemap: provided data are in 1 continuous chunks, the largest of which
has 5957 samples (153.729 s)
smf_iteratemap: map-making requires 1626 MiB (map=28 MiB model calc=1598 MiB)
smf_iteratemap: Continuous chunk 1 / 1 =========
In almost all cases, the raw data files constituting a SCUBA-2 observation will correspond to a single
continuous stream of data samples, taken at roughly 200 Hz. Sometimes however, this may not be the
case, so
you are now told how many chunks of data were found, and how long they were. Something may be wrong if the input data
contains any breaks.
Next comes information about the amount of memory needed to make the map. If insufficient memory is available to
process all the input data together, it will be split into chunks, and a separate map made from each chunk.
These maps are later co-added to form the final output map. This can often result in a poorer map—see Data
Chunking).
Finally, a loop is entered to process each chunk in turn, and you are told which chunk is currently being processed. In this
case there is only one chunk (which is good).
smf_iteratemap: Iteration 1 / 40 ---------------
We now start the first iteration of the iterative algorithm described in Section 3.2, to create a map from the current chunk
of input data. We will be performing —- at most—40 iterations, as set by numiter.
--- Size of the entire data array ------------------------------------------
bolos : 5120
tslices: 5957(2.6 min)
Total samples: 30499840
\begin{terminalv}
--- Quality flagging statistics --------------------------------------------
BADDA: 10805998 (35.43%), 1814 bolos
BADBOL: 10865568 (35.62%), 1824 bolos
DCJUMP: 19631 ( 0.06%),
STAT: 71680 ( 0.24%), 14 tslices
NOISE: 41699 ( 0.14%), 7 bolos
Total samples available for map: 19586411, 64.22% of max (3287.97 bolos)
Now we have a number of statistics describing the cleaned data prior to the first
iteration.
Each sub-array contains a grid of
bolometers, making 5120 bolometers in total over all four sub-arrays. The number of time slices in the concatenated data
after down-sampling is reported. In this case, 5957 time slices over 2.6 minutes, equating to a down-sampled frequency of
about 38 Hz. The total number of samples is the product of the number of bolometers and the number of time slices.
Each of the following lines indicates the percentage of the data that has been flagged as unusable for various
reasons:
- BADDA – flagged as unusable during data acquisition.
- DCJUMP – flagged as unusable because of a sudden step change in the base-line.
- STAT – flagged as unusable because the telescope was stationary (or at least moving too slowly).
- NOISE – flagged as unusable because they were too noisy.
The BADBOL item gives the fraction of bolometers that have been flagged as entirely bad for any of these reasons, and from
which no data will be used.
smf_iteratemap: Calculate time-stream model components
smf_iteratemap: Rebin residual to estimate MAP
smf_iteratemap: Calculate ast
Indicates the models that are being calculated. More information about each individual model is displayed if you set
MSG_FILTER=verbose on the makemap command line.
--- Quality flagging statistics --------------------------------------------
BADDA: 10805998 (35.43%), 1814 bolos ,change 0 (+0.00%)
BADBOL: 11008536 (36.09%), 1848 bolos ,change 142968 (+1.32%)
DCJUMP: 19631 ( 0.06%), ,change 0 (+0.00%)
STAT: 71680 ( 0.24%), 14 tslices,change 0 (+0.00%)
COM: 372786 ( 1.22%), ,change 372786 (+0.00%)
NOISE: 41699 ( 0.14%), 7 bolos ,change 0 (+0.00%)
Total samples available for map: 19214687, 63.00% of max (3225.56 bolos)
Change from last report: -371724, -1.90% of previous
smf_iteratemap: Will calculate chi^2 next iteration
smf_iteratemap: *** NORMALIZED MAP CHANGE: 2.22778 (mean) 60.5292 (max)
More statistics are displayed once the first iteration is completed and the first estimate of the science map has been
generated. Data samples may be flagged as bad within the iterative stage for various reasons, and so these statistics may be
different to those shown prior to the start of the iterative stage. Most significantly, the COM: line shows the percentage of
data that has been rejected because the time stream did not resemble the common-mode signal closely enough. The
“NORMALIZED MAP CHANGE” values are of no real significance on the first iteration since there is no previous map with
which to compare the new map (in fact the numerical values are generated by comparing the new map with a map full of
zeros).
smf_iteratemap: Iteration 2 / 40 ---------------
smf_iteratemap: Calculate time-stream model components
smf_calcmodel_noi: Calculating a NOI variance for each box of 581 samples
using variance of neighbouring residuals.
smf_iteratemap: Rebin residual to estimate MAP
smf_iteratemap: Calculate ast
--- Quality flagging statistics --------------------------------------------
BADDA: 10805998 (35.43%), 1814 bolos ,change 0 (+0.00%)
BADBOL: 11038321 (36.19%), 1853 bolos ,change 29785 (+0.27%)
SPIKE: 36 ( 0.00%), ,change 36 (+0.00%)
DCJUMP: 19631 ( 0.06%), ,change 0 (+0.00%)
STAT: 71680 ( 0.24%), 14 tslices,change 0 (+0.00%)
COM: 393125 ( 1.29%), ,change 20339 (+5.46%)
NOISE: 41699 ( 0.14%), 7 bolos ,change 0 (+0.00%)
Total samples available for map: 19194401, 62.93% of max (3222.16 bolos)
Change from last report: -20286, -0.11% of previous
smf_iteratemap: *** CHISQUARED = 0.997314089857974
smf_iteratemap: *** NORMALIZED MAP CHANGE: 1.78691 (mean) 8.09056 (max)
The second iteration starts, and proceeds in much the same way as the first iteration. The main difference is that the NOI
model is generated at the start of the second iteration. This model measures the variance within each bolometer
time-stream. It is used to weight the bolometer samples when forming the mean sample value in each map
pixel.
NOI is calculated from the residuals left after removal of all other models, and is only calculated once—subsequent
iterations re-use the same NOI values.
The SPIKE: item that has appeared in the quality flagging statistics records the number of samples that have been
rejected as transient spikes in the time-series. This flagging is done by comparing the bolometer values that
fall in each map pixel, and flagging any that appear to be statistical outliers—see configuration parameter
ast.mapspike.
The mean “NORMALIZED MAP CHANGE” value should drop with each subsequent iteration (note, the “max” normalised
map change value can usually be ignored as it records the maximum normalised change in any single map pixel and is thus
highly subject to random variations).
smf_iteratemap: Iteration 3 / 40 ---------------
smf_iteratemap: Calculate time-stream model components
smf_iteratemap: Rebin residual to estimate MAP
smf_iteratemap: Calculate ast
--- Quality flagging statistics --------------------------------------------
BADDA: 10805998 (35.43%), 1814 bolos ,change 0 (+0.00%)
BADBOL: 11050235 (36.23%), 1855 bolos ,change 11914 (+0.11%)
SPIKE: 36 ( 0.00%), ,change 0 (+0.00%)
DCJUMP: 19631 ( 0.06%), ,change 0 (+0.00%)
STAT: 71680 ( 0.24%), 14 tslices,change 0 (+0.00%)
COM: 401600 ( 1.32%), ,change 8475 (+2.16%)
NOISE: 41699 ( 0.14%), 7 bolos ,change 0 (+0.00%)
Total samples available for map: 19185947, 62.91% of max (3220.74 bolos)
Change from last report: -8454, -0.04% of previous
smf_iteratemap: *** CHISQUARED = 0.976599086972009
smf_iteratemap: *** change: -0.0207150028859653
smf_iteratemap: *** NORMALIZED MAP CHANGE: 0.305 (mean) 1.24407 (max)
The mean normalised map change continues to drop with the third iteration, as expected. The total number of samples
going into the map is dropping with each iteration, but only very slowly. This is mainly due to the increased number of
samples being flagged by the COM model, but is at an acceptably small level.
smf_iteratemap: Iteration 4 / 40 ---------------
smf_iteratemap: Calculate time-stream model components
smf_iteratemap: Rebin residual to estimate MAP
smf_iteratemap: Calculate ast
--- Quality flagging statistics --------------------------------------------
BADDA: 10805998 (35.43%), 1814 bolos ,change 0 (+0.00%)
BADBOL: 11056192 (36.25%), 1856 bolos ,change 5957 (+0.05%)
SPIKE: 36 ( 0.00%), ,change 0 (+0.00%)
DCJUMP: 19631 ( 0.06%), ,change 0 (+0.00%)
STAT: 71680 ( 0.24%), 14 tslices,change 0 (+0.00%)
COM: 409883 ( 1.34%), ,change 8283 (+2.06%)
NOISE: 41699 ( 0.14%), 7 bolos ,change 0 (+0.00%)
Total samples available for map: 19177684, 62.88% of max (3219.35 bolos)
Change from last report: -8263, -0.04% of previous
smf_iteratemap: *** CHISQUARED = 0.96404406922676
smf_iteratemap: *** change: -0.0125550177452489
smf_iteratemap: *** NORMALIZED MAP CHANGE: 0.0401588 (mean) 0.151877 (max)
After the fourth iteration the mean normalised map change has dropped to 0.0401588, which is below the
value of 0.05 provided for the maptol parameter by the dimmconfig_bright_compact configuration
file.
Consequently, makemap considers the map to have converged. However, in view of the fact that the
dimmconfig_bright_compact configuration file uses “AST masking” (see Section 3.5.1), it is necessary to do one final
iteration in order to assign correct values to the pixels that lie outside the source mask. Note, when AST masking
is being used, the reported normalised map change values only include pixels that are within the source
mask.
smf_iteratemap: Iteration 5 / 40 ---------------
smf_iteratemap: Calculate time-stream model components
smf_iteratemap: Rebin residual to estimate MAP
smf_iteratemap: Calculate ast
--- Quality flagging statistics --------------------------------------------
BADDA: 10805998 (35.43%), 1814 bolos ,change 0 (+0.00%)
BADBOL: 11068106 (36.29%), 1858 bolos ,change 11914 (+0.11%)
SPIKE: 36 ( 0.00%), ,change 0 (+0.00%)
DCJUMP: 19631 ( 0.06%), ,change 0 (+0.00%)
STAT: 71680 ( 0.24%), 14 tslices,change 0 (+0.00%)
COM: 414727 ( 1.36%), ,change 4844 (+1.18%)
NOISE: 41699 ( 0.14%), 7 bolos ,change 0 (+0.00%)
Total samples available for map: 19172853, 62.86% of max (3218.54 bolos)
Change from last report: -4831, -0.03% of previous
smf_iteratemap: *** CHISQUARED = 0.964032127175568
smf_iteratemap: *** change: -1.19420511923707e-05
smf_iteratemap: *** NORMALIZED MAP CHANGE: 0.0157131 (mean) 0.103135 (max)
smf_iteratemap: ****** Completed in 5 iterations
smf_iteratemap: ****** Solution CONVERGED
Setting 24282 map pixels bad because they contain fewer than 4 samples (=0.01
of the mean samples per pixel).
Total samples available from all chunks: 19172853 (3218.54 bolos)
After the extra iteration required by AST masking has been performed, the final output map is created. It is
always advisable to check the final number of samples available for the map, as a low value will cause your
map to have high noise levels. In this case, 62.86% of the samples are available for the map, which is quite
acceptable—only a couple of percent of the samples have been rejected by the map-making, mostly flagged by the COM
model. The bulk of the bad samples (35.43%) were rejected during data acquisition due to dead bolometers
etc.
Note, the numiter parameter is set to -40 by dimmconfig_bright_compact, meaning that no more than 40
iterations will be performed. In this particular case we only needed 4 iterations (plus a mandatory extra
iteration) to achieve our requested maptol value. But it is possible for some observations—particularly
observations of extended sources—to require more than 40 iterations to converge to a maptol of
0.05. In such cases the screen output will end with a message saying that the solution “FAILED TO
CONVERGE”.
In such cases, you could simply increase numiter and re-run makemap, but you may also want to investigate the cause of
the slow convergence using one or more of the techniques described in Chapter 9, as it may indicate some issue with the
raw data.
Map pixels that receive a very small number of samples are automatically set bad. These are usually the pixels around the
periphery of the observation, and will have very unreliable variance estimates. The threshold is determined by the
hitslimit parameter, which defaults to 1 percent of the mean number of hits per pixel, corresponding to 4 samples per
pixel in this case.
The observation used in this particular case was a short observation of a calibrator, lasting only 2.6 minutes.
Consequently there was no need to divide the data up into multiple chunks in order to fit it into memory.
However, for very long observations, or for shorter observations when using a computer with less than the
recommended amount of memory (see Section 1.2), it may be necessary to process the raw data in multiple chunks.
If this happens, a map is created from each chunk in turn, and all these maps are then added together to
form the final map. The final message “Total samples available from all chunks: 19172853
(3218.54 bolos)” indicates the total number of samples (and equivalent number of bolometers) used from all
chunks. In this case there was only one chunk, so these values are equal to the numbers reported at the end
of the last iteration. Chunking often results in a poorer map and should be avoided if possible—see Data
Chunking).
5.3 Interacting with makemap during a long run
For long observations, makemap can take several hours to run, particularly on slow or low-memory
computers. For this reason it is useful to be able to monitor progress so that potential problems can be detected
early, in order to avoid wasting time.
5.3.1 Monitoring screen output
The most straight-forward way of monitoring progress is to check the values written to the screen by
makemap at the end of each iteration (see the previous section). For instance, if you run makemap as
follows:
% makemap in=^infiles out=outmap config=^conf | tee makemap.log
then the messages displayed by makemap on the screen will also be written to text file makemap.log. This
makes it easy to search the log file whilst makemap is still running, for instance from another terminal
window:
% grep NORMALIZED makemap.log
smf_iteratemap: *** NORMALIZED MAP CHANGE: 1.02829 (mean) 20.7922 (max)
smf_iteratemap: *** NORMALIZED MAP CHANGE: 0.739832 (mean) 9.81836 (max)
smf_iteratemap: *** NORMALIZED MAP CHANGE: 0.370128 (mean) 5.42933 (max)
smf_iteratemap: *** NORMALIZED MAP CHANGE: 0.258304 (mean) 3.2717 (max)
smf_iteratemap: *** NORMALIZED MAP CHANGE: 0.205415 (mean) 2.66815 (max)
smf_iteratemap: *** NORMALIZED MAP CHANGE: 0.177044 (mean) 2.28417 (max)
smf_iteratemap: *** NORMALIZED MAP CHANGE: 0.159569 (mean) 2.05803 (max)
This displays the normalised change in maps between successive iterations, for the iterations that have so far
been completed. If the mean normalised map change is not decreasing smoothly, (for instance if it is oscillating
around a fixed value), then there is a potential problem. In which case you may want to interrupt
the makemap process, rather than waiting potentially for several hours just to end up with a bad
map.
Likewise, it is useful to monitor the total number of samples that are being pasted into the map at the end of
each iteration:
% grep "Total samples" makemap.log
Total samples: 299919360
Total samples available for map: 174105275, 58.05% of max (2972.2 bolos)
Total samples available for map: 171284110, 57.11% of max (2924.03 bolos)
Total samples available for map: 171242020, 57.10% of max (2923.32 bolos)
Total samples available for map: 171239147, 57.10% of max (2923.27 bolos)
The lower the percentage of samples included in the map, the greater will be the noise in the map. If this
number drops much below 50% then you may want to think about aborting makemap and investigating why the
number is so low (e.g. by looking at the “quality statistics” displayed at the end of each iteration, to determine
the cause of the data loss).
Likewise, information can be gathered about chunking:
% grep chunk makemap.log
smf_iteratemap: provided data are in 7 continuous chunks, the largest of
which has 487 samples (10.1534 s)
smf_iteratemap: Continuous chunk 1 / 7 =========
smf_iteratemap: Adding map estimated from this continuous chunk to total
smf_iteratemap: Continuous chunk 2 / 7 =========
smf_iteratemap: Adding map estimated from this continuous chunk to total
smf_iteratemap: Continuous chunk 3 / 7 =========
smf_iteratemap: Adding map estimated from this continuous chunk to total
This indicates that the raw data has gaps in it, resulting in seven maps being made, one from each continuous
chunk, before the final map is made by combining all the individual maps. Chunking can produce sub-optimal
maps, and should normally be investigated—see Data Chunking.
5.3.2 Monitoring the map at the end of each iteration
The map created at the end of each iteration is normally discarded, but can be saved by setting configuration
parameter itermap to either 1 or 2. By default, these “itermaps” are written to an extension of the main output
NDF as described in Section 6.2, and cannot be viewed until makemap has completed. However, an alternative
destination for these itermaps can be specified on the makemap command-line as follows, allowing them to be
viewed whilst makemap is still running:
% cat conf
^STARLINK_DIR/share/smurf/dimmconfig_jsa_generic.lis
itermap=1
%
% makemap in=^infiles out=outmap config=^conf itermaps=myitermaps
Note the distinction between the “itermaps” (plural) command-line option, and the “itermap” (singular)
configuration parameter. As soon as the first iteration has finished, the above command will create a
new file called myitermaps.sdf in which each itermap will be stored as soon as it is created. The
file is closed after each itermap is written, and re-opened again before writing the next itermap.
This is an example of a container file, where a single .sdf file contains several NDFs, each holding
a different map (see Section 1.3.1). You can list the maps in such a file using Kappa ndfecho as
follows:
% ndfecho myitermaps
myitermaps.CH00I001
myitermaps.CH00I002
myitermaps.CH00I003
myitermaps.CH00I004
myitermaps.CH00I005
myitermaps.CH00I006
myitermaps.CH00I007
...
The name of each itermap is of the form “CHxxIyyy”, where “xx” is the chunk number and “yyy” is the
iteration number. In the common case where all raw data are processed in a single chunk, “xx” will be “00” for
all itermaps.
You can view a single itermap using:
% gaia myitermaps.CH00I004
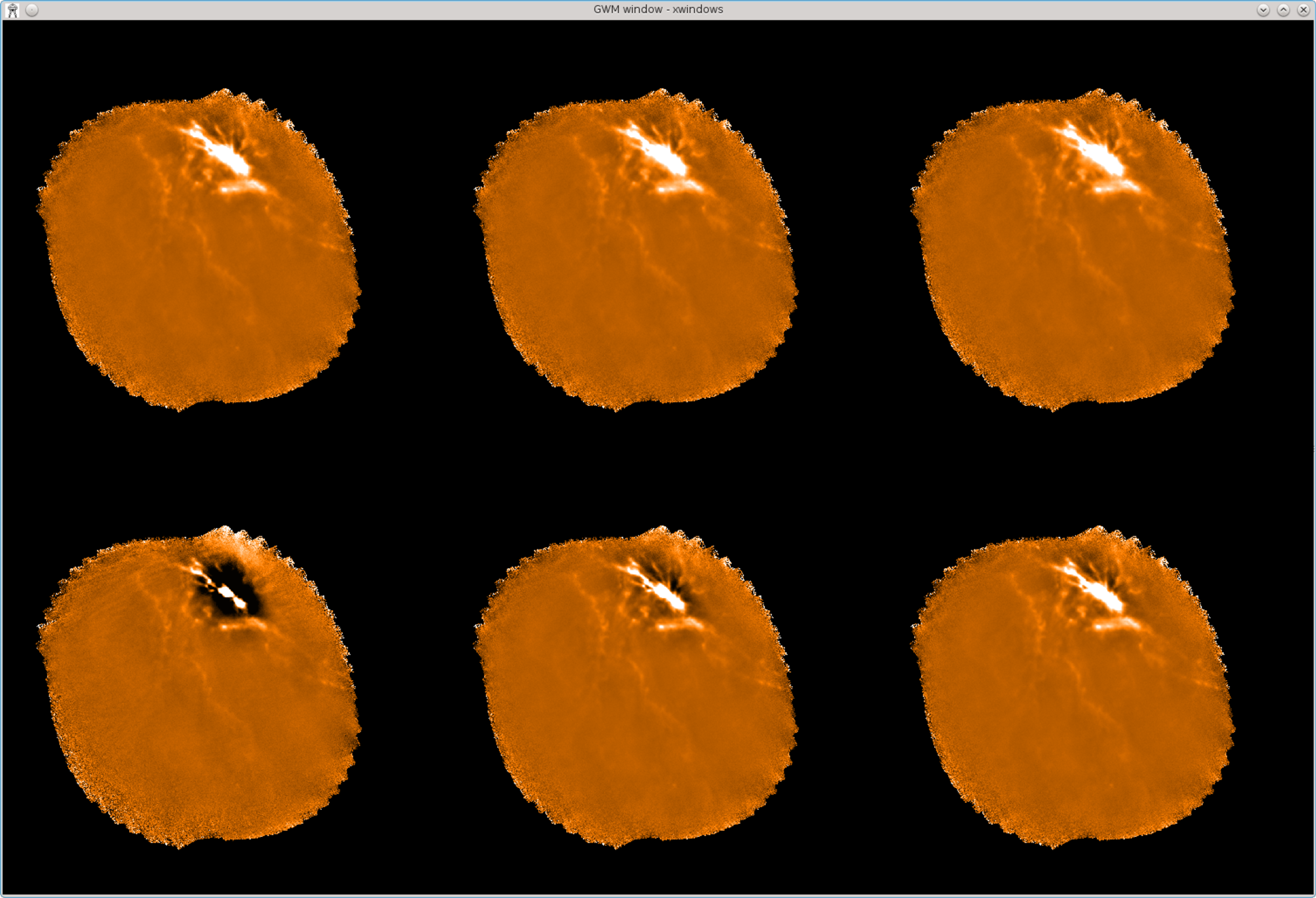
If you want to view several itermaps side-by-side, you can use Kappa picgrid to divide the
screen up into a grid of “pictures”, then use picsel to pick each picture in turn, and use display
to display an itermap in each one. For instance to display the first six itermaps side-by-side in a
grid, all
with the same scaling, without any axes, do:
% gdclear
% picgrid 3 2
% picsel 1
% display myitermaps.CH00I001 axes=no mode=percentiles percentiles=\[2,98\]
% picsel 2
% display myitermaps.CH00I002 mode=current
% picsel 3
% display myitermaps.CH00I003 mode=current
% picsel 4
% display myitermaps.CH00I004 mode=current
% picsel 5
% display myitermaps.CH00I005 mode=current
% picsel 6
% display myitermaps.CH00I006 mode=current
This produces the display shown in Figure 5.1. Kappa display has many options for controlling things like data
scaling, axis annotation and style, colour table, graphics device, etc..
Alternatively, you can combine the itermaps into a 3D cube, and then use the
cube-viewing facilities of Gaia to scroll through the itermaps in the style of a movie (see
Section 6.2):
% paste in=myitermaps out=itercube shift=\[0,0,1\]
% gaia itercube
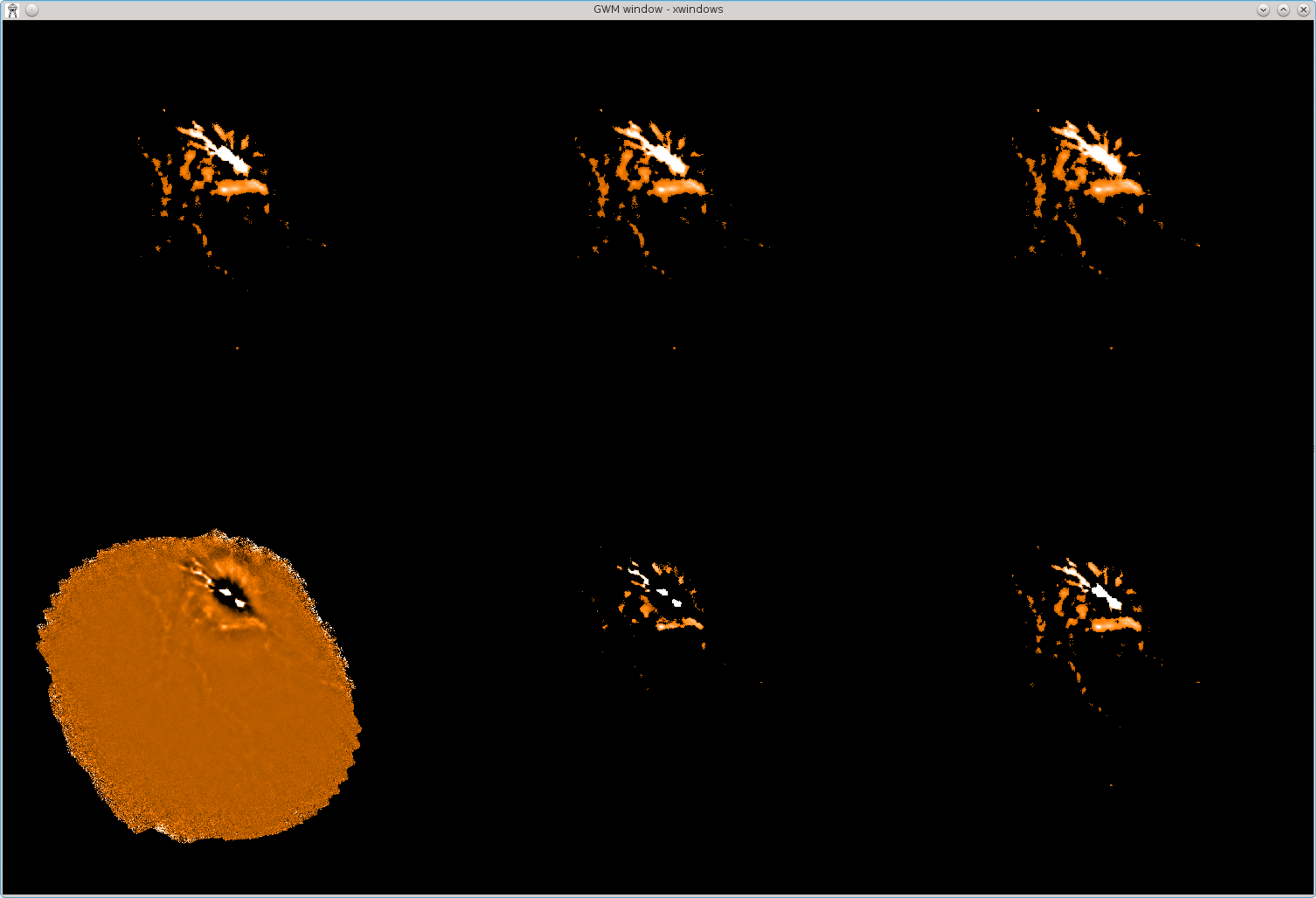
It is sometimes informative to look at the change between itermaps, to see the incremental effect of each
iteration. This is most easily done if the itermaps are first stacked into a cube as described above. The next step
is to produce a copy of the cube in which the pixel origin is moved by one pixel along the third axis (i.e. the axis
that enumerates iteration). Finally subtract one cube from the other, and display the resulting difference cube
(this example shows how command-line parameters can often be specified by position instead of by name when
running Kappa commands—check the help information for each command to see the order in which options
should be supplied):
% paste myitermaps out=itercube shift=\[0,0,1\]
% slide itercube itercube-shifted \[0,0,1\] near
% sub itercube-shifted itercube diffcube
%
% picsel 1
% display diffcube’(,,1)’ axes=no mode=percentiles percentiles=\[2,98\]
% picsel 2
% display diffcube’(,,2)’ mode=current
% picsel 3
% display diffcube’(,,3)’ mode=current
% picsel 4
% display diffcube’(,,4)’ mode=current
% picsel 5
% display diffcube’(,,5)’ mode=current
% picsel 6
% display diffcube’(,,6)’ mode=current
This produces the display shown in Figure 5.2.
Viewing the mask after each iteration:
The above plots are based on the itermaps that are created by adding “itermaps=1”
to your makemap configuration. You can instead use “itermaps=2”, which causes each
itermap to be masked so that pixels outside the current source mask are set bad (i.e.
blank).
Figure 5.3 shows the resulting itermaps.
The main point of this section is to emphasise that all the above investigations can be performed whilst makemap
is still running, if you specify an explicit file for the itermaps using the “itermaps command-line option.
This allows you to decide if the reduction is progressing as expected, and whether to interrupt the
reduction.
5.3.3 Interrupting makemap
If you interrupt makemap by pressing control-C on the keyboard (or equivalently by sending an INT signal to
the makemap process), it will continue until the current iteration is completed and then will issue
a prompt with options allowing you to save the current map before closing, as in the following
example:
% makemap in=^infiles out=outmap config=^conf itermaps=myitermaps
Out of 29 input files, 1 was a dark, 1 was a fast flat and 27 were science
Processing data from instrument ’SCUBA-2’ for object ’OMC1 tile10’ from the
following observation :
...
...
<press control-C>
...
...
--- Quality flagging statistics
--------------------------------------------
BADDA: 25525734 (20.86%), 267 bolos ,change 0 (+0.00%)
BADBOL: 31261854 (25.55%), 327 bolos ,change 0 (+0.00%)
SPIKE: 339 ( 0.00%), ,change 4 (+1.19%)
DCJUMP: 146703 ( 0.12%), ,change 0 (+0.00%)
STAT: 160000 ( 0.13%), 125 tslices,change 0 (+0.00%)
COM: 1827454 ( 1.49%), ,change 3000 (+0.16%)
NOISE: 5640518 ( 4.61%), 59 bolos ,change 0 (+0.00%)
Total samples available for map: 89135553, 72.84% of max (932.361 bolos)
Change from last report: -3001, -0.00% of previous
smf_iteratemap: *** CHISQUARED = 0.998314983615372
smf_iteratemap: *** change: -0.0014711436109287
smf_iteratemap: *** NORMALIZED MAP CHANGE: 0.00941492 (mean) 2.9521 (max)
>>>> Interrupt detected!!! What should we do now? Options are:
1 - abort immediately with an error status
2 - close the application returning the current output map
3 - do one more iteration to finalise the map and then close
NOTE - another interrupt will abort the application, potentially leaving
files in an unclean state.
INTOPTION - What to do now (1-3) /3/ >
At this point you should respond to the prompt for parameter INTOPTION by typing 1, 2 or 3 followed by
<return>. Option 1 aborts the process with an error status. Option 2 causes makemap to tidy up its internals and
create a map from the current models just as if the iterative process had reached convergence. Option 3 causes
one further iteration to be performed, without masking before creating a final map (you should use this option
if you have been using AST masking—see Section 3.5.1).
Tip: Note if you are running
makemap from within a script, then the handling of control-C interrupts will
probably be quite different, because the shell process (or perl, python or whatever) will catch the interrupt
before it gets to the
makemap process. This usually causes the shell process to die but leaves the
makemap process running in the background. Instead of pressing control-C on the keyboard, you
should find the process ID for the
makemap process itself, and then send an INT signal to that process
explicitly:
% ps aux | grep makemap
dsb 25407 0.0 0.0 43716 1952 pts/5 0:00 makemap
% kill -s INT 25407
5.4 Tips and tricks
5.4.1 Aligning your map with a pre-existing image
If you want to compare SCUBA-2 data with a map of the same region taken with a
different instrument, you will usually want to create the SCUBA-2 map using the same
pixel grid as the other map so that you can compare pixel values directly in the two
maps.
You can use the REF command-line parameter to do this. For instance, if your other map is in file herschel.sdf,
you can do:
% makemap in=^infiles out=outmap config=^conf ref=herschel
This will cause the output map in file outmap.sdf to use the same pixel grid as herschel.sdf.
This means that a given point on the sky will have the same pixel coordinates in both maps, and so
for instance you could divide one by the other to get a ratio map without any further alignment
step:
% div outmap herschel ratio_map
This will divide outmap.sdf by herschel.sdf and put the ratio in
ratio_map.sdf.
However, by default makemap will trim the output map to exclude blank borders, thus the two maps may have
different dimensions. NDF-based applications such as Kappa div automatically take account of this when
comparing corresponding pixels in two NDFs—see Figure 5.4.
If you want to force the output map to have certain bounds in pixel coordinates, you can do so by assigning
values to the LBND and UBND parameters when running makemap. For instance, if you want to force the output
map to have the same dimensions as the reference map, you could use Kappa ndftrace to list the properties of
the reference NDF, including its pixel bounds, and then assign these bounds to LBND and UBND parameters when
running makemap:
% ndftrace herschel
NDF structure /home/dberry/herschel1:
Title: Example Herschel data
Shape:
No. of dimensions: 2
Dimension size(s): 351 x 400
Pixel bounds : -50:300, 1:400
Total pixels : 140400
...
...
% makemap in=^infiles out=outmap config=^conf ref=herschel \
lbnd=\[-50,1\] ubnd=\[300,400\]
The resulting map in outmap.sdf will have the same pixel bounds as hershel.sdf.
5.4.2 Limiting the amount of memory used by makemap
There may be occasions when you need to limit the amount of memory used by makemap–for instance to leave
enough memory for other processes to function correctly. For machines with more than 20 GB of memory, the
default is to leave 4 GB free for other processes. For machines with less than than 20 GB of memory, the default
is to leave 20% of the total memory free. This default can be over-ridden by indicating the maximum amount of
memory that makemap should use by setting a value for the command-line parameter MAXMEM. For
instance:
% makemap in=^infiles out=outmap config=^conf maxmem=50000
will ensure that makemap never uses more than 50000
MiB
of memory. Be aware though that for larger observations this may cause the data to be processed in multiple
chunks rather than a single continuous chunk, resulting in a poorer map.
5.4.3 Re-using previously cleaned data to speed up map-making
Usually, you will create your SCUBA-2 maps using one of the standard configuration files distributed with
smurf (see Section 3.7). However, for certain problematic observations it may sometimes be necessary to
experiment with several different configurations to find one that produces an acceptable map. In such cases a
significant amount of time can be saved by re-using pre-cleaned raw data each time you make a map, rather
than re-cleaning it each time.
By default makemap assumes that the supplied data are raw data that require cleaning before being used to
make a map. However, if you include “doclean = 0” in your configuration, then makemap will skip the entire
cleaning process and move directly on to the iterative map-making stage, thus saving significant
time.
Obviously though, in this case you need to make sure that the data you supply to makemap have already been
cleaned. There are two ways to get cleaned data:
-
(1)
- You can run
makemap initially on the un-cleaned data (i.e. the original raw data), and force makemap
to dump the cleaned data to disk before going onto the iterative map-making stage. You can then
re-use this cleaned data in subsequent invocations of makemap. To dump the cleaned data, you
need to add “exportclean = 1” to the configuration on your initial run of makemap—don’t forget
to remove it for subsequent runs! The cleaned data are put into files in the current directory, one for
each sub-array, with names such as s8c20120706_00037_0003_con_res_cln.sdf. Note the _cln
on the end of the name indicating cleaned data. The base name, “s8c20120706_00037_0003” in this
case, is taken from the first raw data file that contributes to each chunk. If the entire observation
is processed in one chunk (as it should usually be) then there will be one such _cln file for each
sub-array. If the observation is split into multiple chunks, then there will be multiple _cln files for
each sub-array (one for each chunk).
-
(2)
- You can use the separate Smurf task sc2clean to clean the data as described in Appendix A.
Copyright © 2014-2021 Science and Technology Facilities Council,
East Asian Observatory