Processing math: 100%
3 Starlink
3.1 MEANARC
-
Purpose:
-
Pastes the average of two arc wavelength scales onto a spectrum.
-
Language:
-
Perl script.
-
Description:
-
A common data reduction task is to apply the wavelength calibration based an arc lamp
spectrum to an object frame or several object frames. It is often the case that we have
two arc lamp exposures which ‘bracket’ the astronomy frames in case there is some time
dependence of the wavelength scale. FIGARO XCOPI allows a weighted mean of the
wavelength scales of two arc frames to be applied to an object frame. This script takes
this one step further by working out the weighting – based upon Modified Julian Dates
extracted from FITS header information which should have propagated into the Starlink
data structures holding the object frames. If FITS headers are not present the script won’t
work as is, but could be modified to obtain date information from a different source.
This is a perl script – don’t be alarmed! If it does what you need then it’s very simple to
use. Perl was chosen as a simple floating point calculation is needed. The script could be
modified to drive a different FIGARO application.
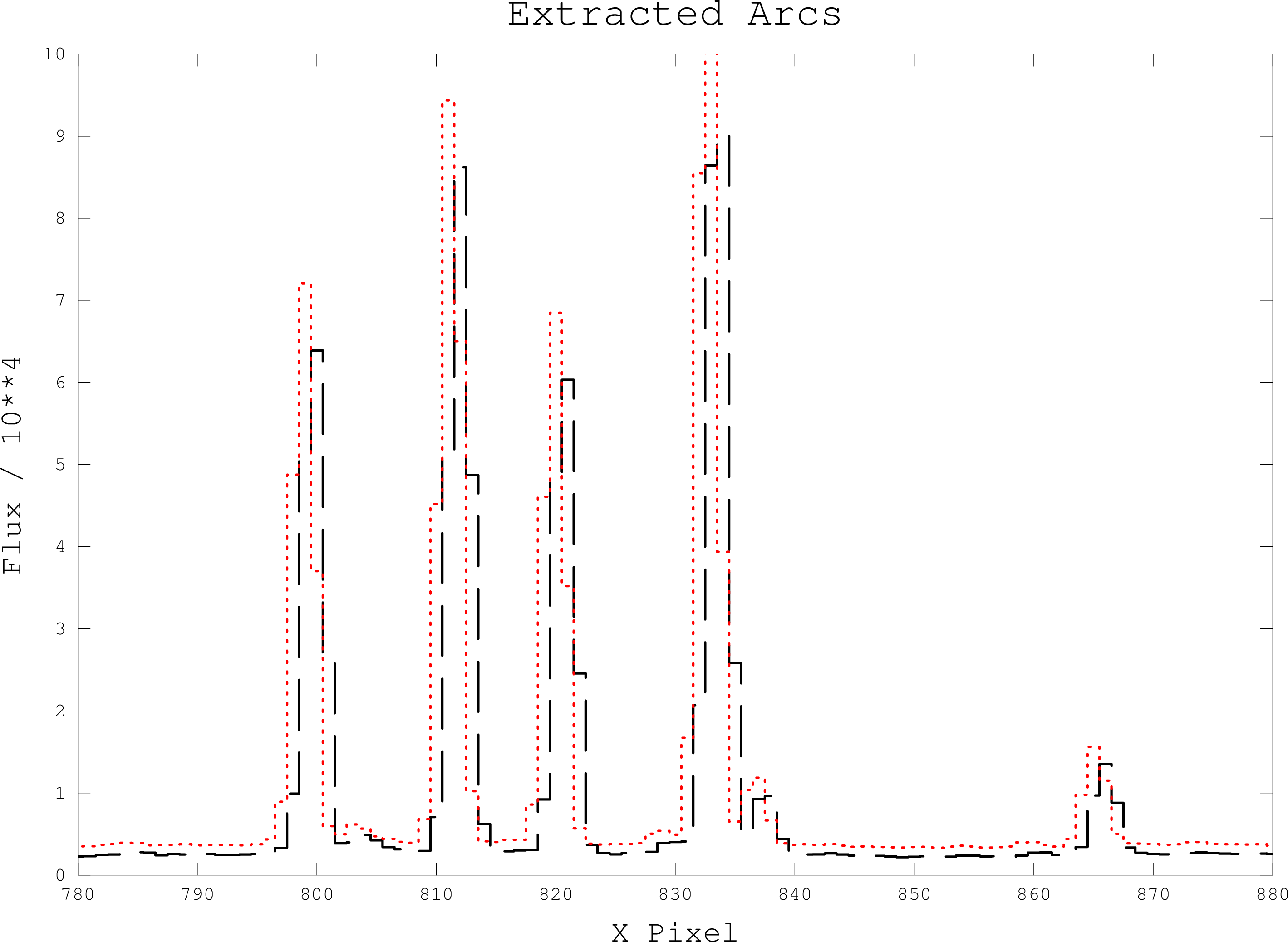
This script will take two wavelength-calibrated arcs, typically bracketing object frames,
use FIGARO XCOPI to find the mean wavelength scale for each of the object
frames and apply this scale to the data. This script is suitable for processing spectra
wavelength-calibrated with FIGARO ECHARC.
HDSTRACE is used to search for the MJD record of a FITS header from which the
Modified Julian Date for the file can be extracted. The weighting of the ‘mean’ wavelength
scale for an object frame is based on the MJD for each of the two arcs and that of the object
itself. Schematically the weighting is:
OUTPUT=ARC1+TOBJ−TARC1TARC2−TARC1×(ARC2−ARC1)
where:
-
- OUTPUT is the wavelength scale produced.
-
- ARC1 is the wavelength scale of ARC1.
-
- ARC2 is the wavelength scale of ARC2.
-
- TARC1
is the MJD for ARC1.
-
- TARC2
is the MJD for ARC2.
-
- TOBJ
is the MJD for the object.
-
Usage:
-
You can simply invoke this script with no arguments and you will be prompted for the required
information. Alternatively, you can supply the arguments on the command line. For example, if
you have two object frames obj1 and obj2 bracketed by the arc exposures arc1 and arc2 this
would be a suitable way to invoke the script:
% meanarc arc1 arc2 obj1 obj2
If supplied, command-line arguments must be in this order:
-
(1)
- First Arc. Name of the first arc frame file.
-
(2)
- Second Arc. Name of the second arc frame file.
-
(3)
- List of object frames. Names of the files to which the calibration should be applied.
You can supply as many names as you like, separated by spaces. If you are prompted
for a list of objects, then you should supply a comma-separated list of object frames.
Missing command-line arguments are prompted for.
-
Source code:
-
In a standard Starlink installation the source code for MEANARC can be found in the
file:
/star/examples/sc3/meanarc
-
Notes:
-
-
(1)
- FIGARO V5.0-0 or higher is required.
-
(2)
- By default, this script modifies the wavelength scales of the object files on which it
acts. To create new files with a postfix (e.g. file_wcal.sdf from file.sdf) modify
the value of the variable $Postfix as commented in the script.
-
(3)
- If FITS header information has not been propagated to the Starlink data structure,
or there is no Modified Julian Date present, this script will not work. The routine
‘getmjd’ in the script could be modified to use date information from a different
source. See the script for details.
3.2 ECHTRACT
-
Purpose:
-
Script to extract orders from a reduced échelle image to individual NDF files suitable for
reading into DIPSO.
-
Language:
-
C shell script.
-
Description:
-
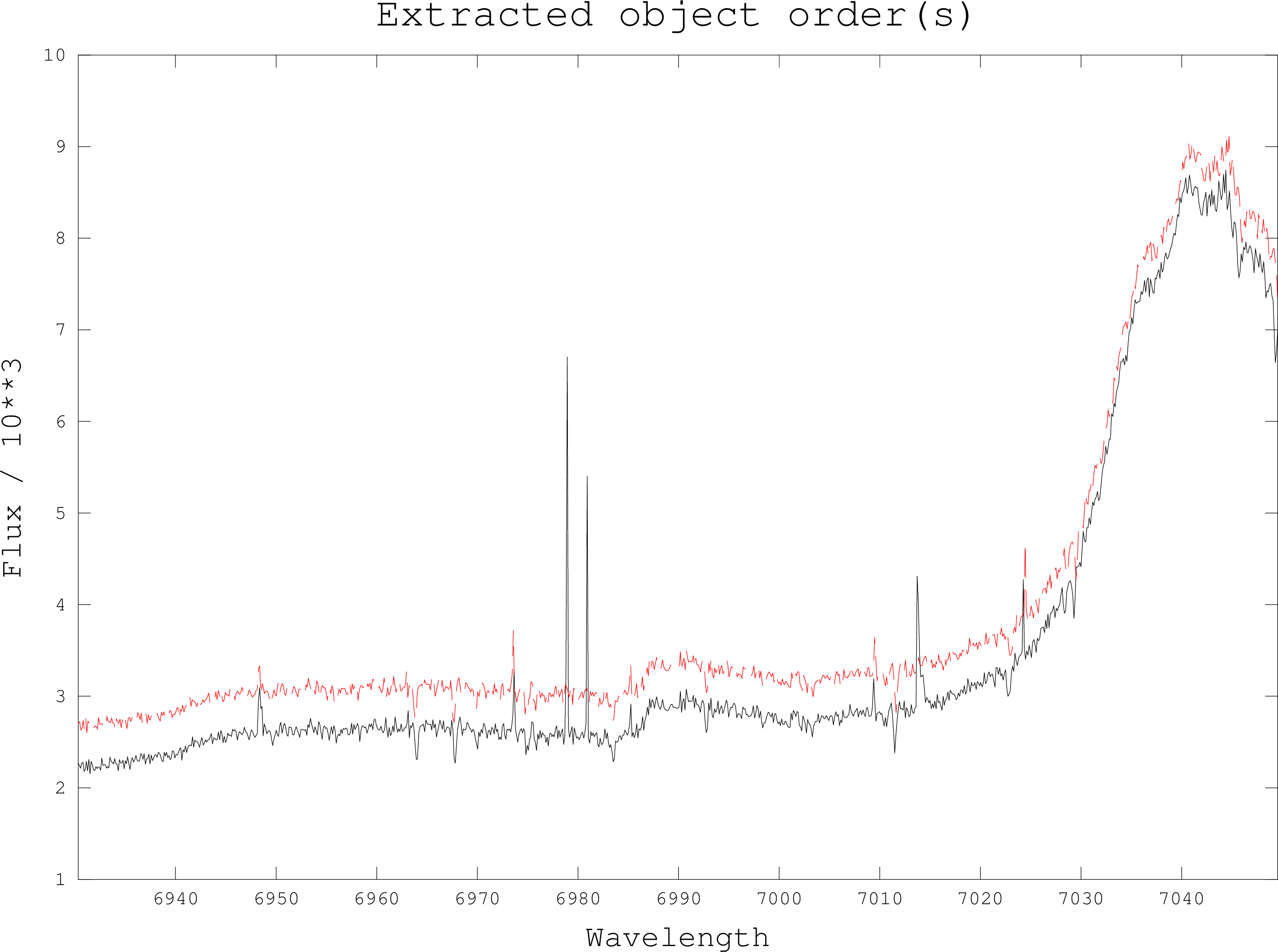
This script performs a common task; slicing out individual orders from a collapsed,
extracted échelle image. In a collapsed échelle image each row of the 2-D image is a
single order from the echellogram. Each order has its own wavelength scale, which is
stored in an NDF extension. This script will pair up each order with its wavelength scale
and output them as individual NDFs with flux data in the main NDF data array and
wavelength data in the AXIS(1) data array, in other words, an NDF which can be read by
DIPSO, FIGARO and so on.
The input image should be one output from ECHOMOP (produced using ech_result
or echmenu Option 14) or FIGARO ECHARC, or one which consists of a 2-dimensional
image where each order occupies a single line of the image.
There are two versions of this script available:
- echtract_echomop is specifically for processing ECHOMOP output.
- echtract_echarc is specifically for processing FIGARO ECHARC output.
The scripts expect the wavelength data in the input images to be stored in NDF extensions as
follows:
- .MORE.ECHELLE.ECH_2DWAVES for ECHOMOP data.
- .AXIS(1).MORE.FIGARO.DATA_ARRAY.DATA for ECHARC data.
if this is not the case you can still use the script by modifying the value of the AXISDATA variable
in the scripts.
The full all-order wavelength scales would normally be propagated to the output NDFs; to
reduce the size of the individual-order NDFs this extension is deleted in each output file. You
can modify this behaviour by commenting out the part of the script which ‘shrinks’ the
per-order NDFs.
The main 2-D order array is expected to be in the NDF main DATA_ARRAY (it will be for
ECHOMOP or ECHARC data). If you want to take the data from a different location, then set the
variable FLUXDATA in the scripts to reflect the location. For example, if the data are in the
extension .MORE.ECHELLE.DATA_ARRAY, then edit the appropriate line in the script
to:
set FLUXDATA = ’.MORE.ECHELLE.DATA_ARRAY’;
Be sure to include the leading ‘.’ or the extension will not be found.
-
Usage:
-
You can simply invoke this script with no arguments and you will be prompted for the required
information. Alternatively, you can supply the arguments on the command line. For example, if
you have a collapsed échelle spectrum extobj.sdf which contains 42 orders and you want
each order to be stored in an NDF called extord_nn.sdf, where nn is the number of the order,
invoke the script thus:
% echtract extobj 1 42 extord
If supplied, command-line arguments must be in this order:
-
(1)
- Input image. Name of the image containing the échelle orders.
-
(2)
- Number of first order. Number of the first échelle order to be extracted.
-
(3)
- Number of last order. Number of the last échelle order to be extracted.
-
(4)
- Output root. Root name for output images, e.g. a value ech will lead to output files
ech_01.sdf, ech_02.sdf and so on.
Missing command-line arguments are prompted for.
-
Source code:
-
In a standard Starlink installation the source code for ECHTRACT can be found in
the:
/star/examples/sc3/echtract_echomop for ECHOMOP data.
/star/examples/sc3/echtract_echarc for FIGARO ECHARC data.
-
Notes:
-
-
(1)
- FIGARO V5.0-0 or higher is required.
-
(2)
- KAPPA V0.9-0 or higher is required.
-
(3)
- By default, wavelength data in the input file should be in the extension:
- .MORE.ECHELLE.ECH_2DWAVES for ECHOMOP data.
- .AXIS(1).MORE.FIGARO.DATA_ARRAY.DATA for ECHARC data.
Use HDSTRACE to check this.
-
(4)
- By default, the flux array is assumed to be in the main
of the input file. Use HDSTRACE to check this.
-
(5)
- This script performs a Starlink login, setup for FIGARO and KAPPA commands. This is so
that the script need not be ‘source’d to use. You can reduce the script set up time (and get
rid of the login/setup messages) if you have already done a Starlink login, setup for
FIGARO and for KAPPA. Edit out the lines as indicated in the script then, to use this script,
you must source it. For example:
% source echtract extobj 1 42 extord
3.3 STACKGEN
-
Purpose:
-
Script to generate a DIPSO stack containing orders from a collapsed échelle image.
-
Language:
-
C shell script.
-
Description:
-
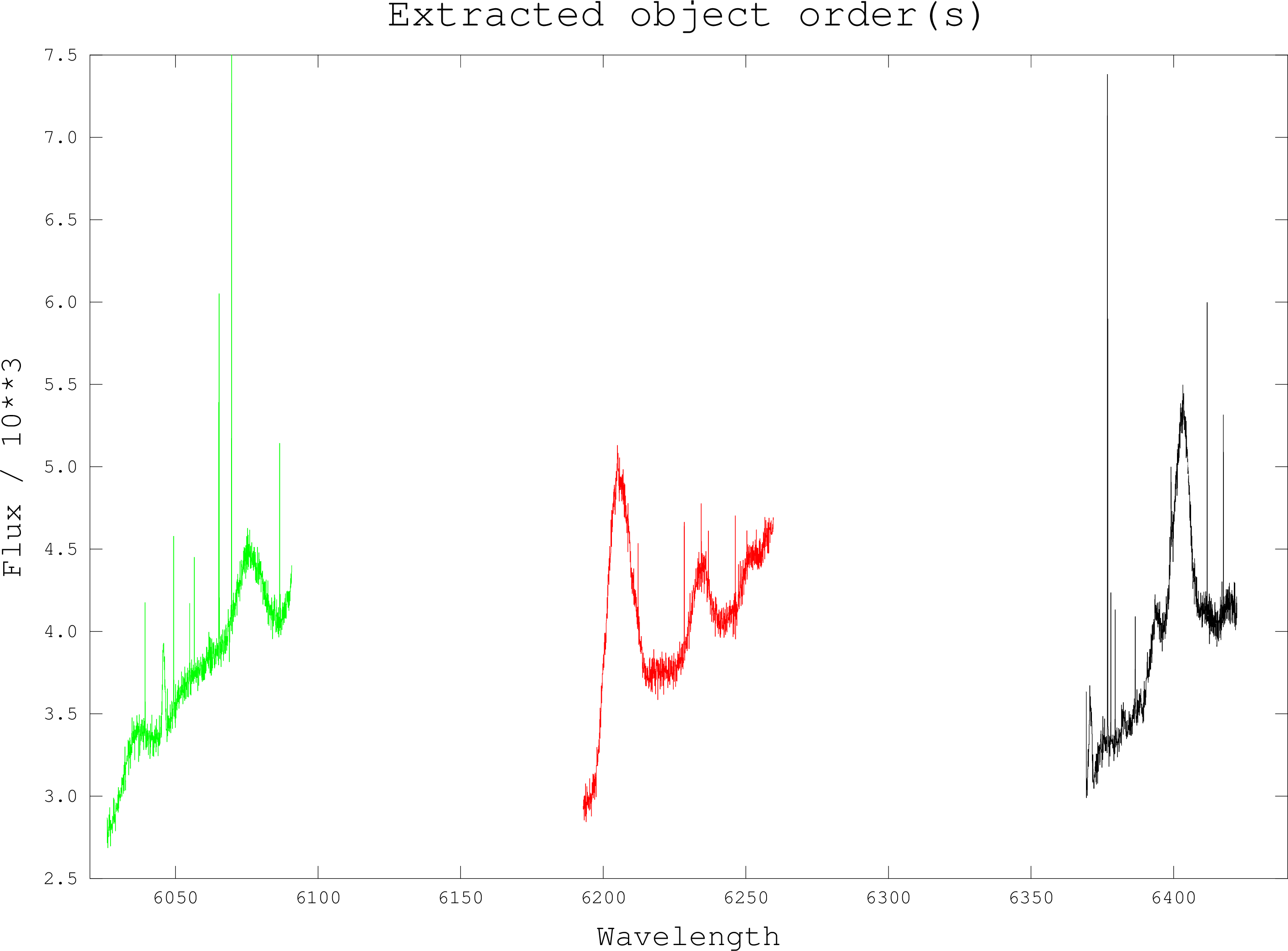
This script converts an NDF containing a collapsed échelle spectrum into a DIPSO stack
where each stack entry holds one order from the echellogram. In a collapsed échelle
image each row of the 2-D image is a single order from the echellogram. Each order has its
own wavelength scale, which is stored in an NDF extension. This script will pair up each
order with its wavelength scale and output them as individual NDFs with flux data in
the main NDF data array and wavelength data in the AXIS(1) data array, in other words,
an NDF which can be read by DIPSO, FIGARO and so on. Once the orders have been
output to their own NDFs, they are read into DIPSO and then saved as a DIPSO stack.
The intermediate NDFs are then deleted.
The input image should be one output from ECHOMOP (produced using ech_result
or echmenu Option 14) or FIGARO ECHARC, or one which consists of a 2-dimensional
image where each order occupies a single line of the image.
There are two versions of this script available:
- stackgen_echomop is specifically for processing ECHOMOP output.
- stackgen_echarc is specifically for processing FIGARO ECHARC output.
The scripts expect the wavelength data in the input images to be stored in NDF extensions as
follows:
- .MORE.ECHELLE.ECH_2DWAVES for ECHOMOP data.
- .AXIS(1).MORE.FIGARO.DATA_ARRAY.DATA for ECHARC data.
if this is not the case you can still use the script by modifying the value of the AXISDATA variable
in the scripts.
The main 2-D order array is expected to be in the NDF main DATA_ARRAY (it will be for
ECHOMOP or ECHARC data). If you want to take the data from a different location, then set the
variable FLUXDATA in the scripts to reflect the location. For example, if the data are in the
extension .MORE.ECHELLE.DATA_ARRAY, then edit the appropriate line in the script
to:
set FLUXDATA = ’.MORE.ECHELLE.DATA_ARRAY’;
Be sure to include the leading ‘.’ or the extension will not be found.
-
Usage:
-
You can simply invoke this script with no arguments and you will be prompted for the required
information. Alternatively, you can supply the arguments on the command line. For example, if
you have a collapsed échelle spectrum extobj.sdf which contains 42 orders and you
want the stack to be called ech_STK.sdf, which you can then read in DIPSO with the
command:
invoke the script thus:
% stackgen extobj 1 42 ech
If supplied, command-line arguments must be in this order:
-
(1)
- Input image. Name of the image containing the échelle orders.
-
(2)
- Number of first order. Number of the first échelle order to be extracted.
-
(3)
- Number of last order. Number of the last échelle order to be extracted.
-
(4)
- Output stack. Root name for output stack, e.g. a value ech will lead to output DIPSO
stack
ech_STK.sdf. This is also used as the root name for the temporary single-order
NDFs.
Missing command-line arguments are prompted for.
-
Source code:
-
In a standard Starlink installation the source code for STACKGEN can be found in
the:
/star/examples/sc3/stackgen_echomop for ECHOMOP data.
/star/examples/sc3/stackgen_echarc for FIGARO ECHARC data.
-
Notes:
-
-
(1)
- FIGARO V5.0-0 or higher is required.
-
(2)
- KAPPA V0.9-0 or higher is required.
-
(3)
- By default, wavelength data in the input file should be in the extension:
- .MORE.ECHELLE.ECH_2DWAVES for ECHOMOP data.
- .AXIS(1).MORE.FIGARO.DATA_ARRAY.DATA for ECHARC data.
Use HDSTRACE to check this.
-
(4)
- By default, the flux array is assumed to be in the main
of the input file. Use HDSTRACE to check this.
-
(5)
- This script performs a Starlink login, setup for FIGARO commands, setup for KAPPA
commands and DIPSO setup. This is so that the script need not be ‘source’d to use. You can
reduce the script set up time (and get rid of the login/setup messages) if you have
already done a Starlink login, setup for FIGARO, KAPPA, and DIPSO. Edit out the
lines as indicated in the script then, to use this script, you must source it. For
example:
% source stackgen extobj 1 42 ech
3.4 TRACEPOLY & ARCPOLY
-
Purpose:
-
TRACEPOLY is a script to extract trace polynomial coefficients from an ECHOMOP
data reduction structure file. ARCPOLY is a similar script which extracts wavelength
polynomial coefficients from an ECHOMOP data reduction structure file.
-
Language:
-
C shell script.
-
Description:
-
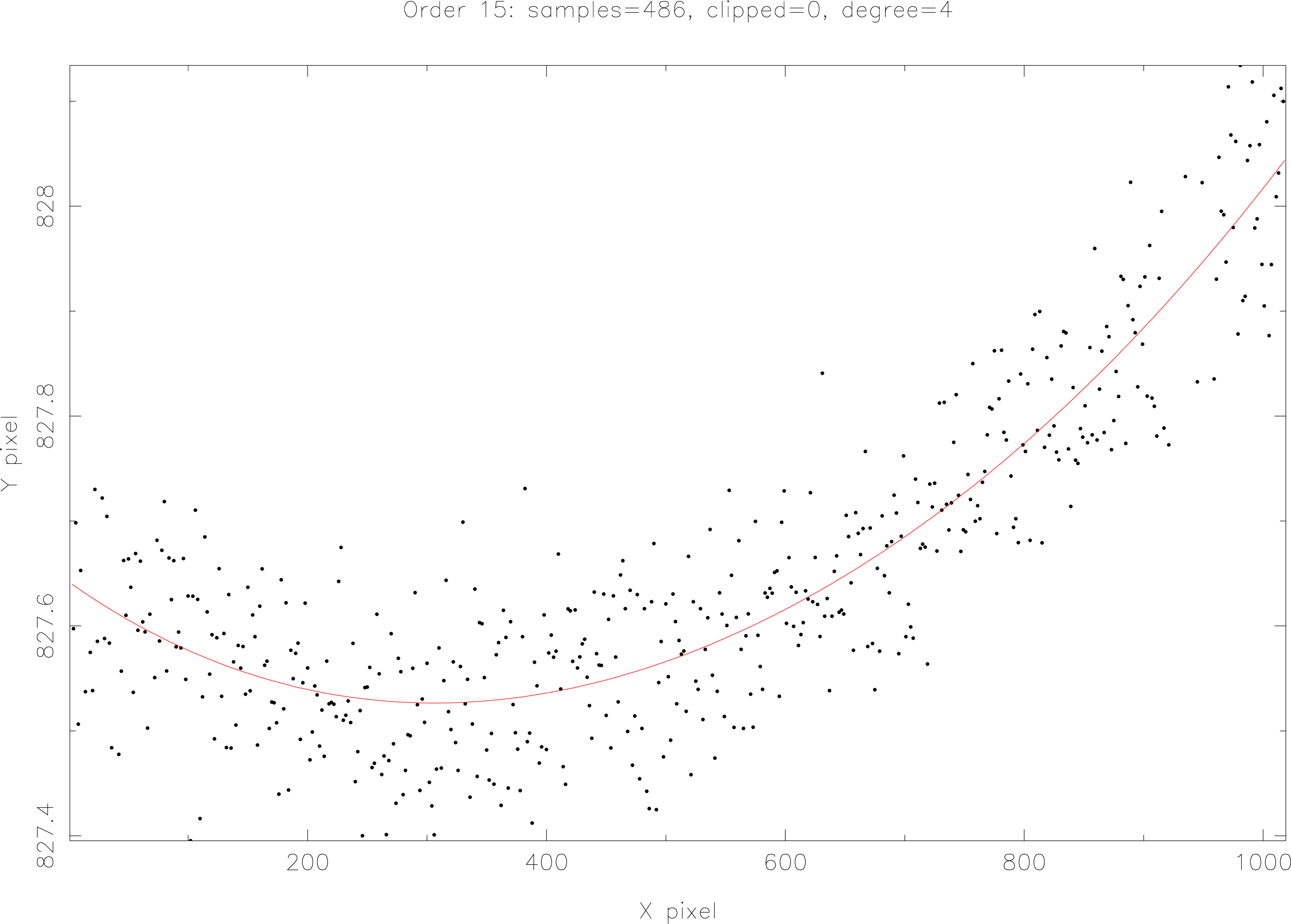
TRACEPOLY uses the Starlink HDSTRACE utility to look for the object
<file>.MORE.ECHELLE.TRC_POLY
in the ECHOMOP reduction structure file <file>.sdf which holds the coefficients of order trace
polynomials as determined by the ECHOMOP task ech_trace. This allows easy access to the
trace polynomials outside of ECHOMOP tasks. The script can be modified to display other
information from ECHOMOP reduction structures. An example of this is ARCPOLY which looks
for the object
<file>.MORE.ECHELLE.W_POLY
which holds the wavelength-scale polynomials for the reduction.
-
Usage:
-
You can simply invoke the scripts with no arguments and you will be prompted for the required
information. Alternatively, you can supply the arguments on the command line. For
example:
will display the first seven polynomial coefficients for order 4 of the data in the reduction
structure file rdf68.sdf.
If supplied, command-line arguments must be in this order:
-
(1)
- Reduction structure. Name of the ECHOMOP reduction structure file.
-
(2)
- Number of the order. Number of the order trace or wavelength scale to be displayed.
-
(3)
- Maximum coefficients. (Default value 8.) Number of polynomial coefficients to be
displayed.
Arguments 1–2 will be prompted for if not given on the command line.
Argument 3 defaults to the value 8 if not given on the command line.
-
Source code:
-
In a standard Starlink installation the source codes for TRACEPOLY and ARCPOLY can be
found in the files:
/star/examples/sc3/tracepoly
and
/star/examples/sc3/arcpoly
3.5 Automated ECHOMOP reductions
This family of scripts can be used to manage the automatic reduction of échelle data based upon a
single template reduction performed manually. The scripts are as follows:
-
PREPRUN1
- Driver script for an automated reduction run. Prompts for details of
the dataset.
-
PREPRUN2
- Driver script for an automated reduction run. Should be edited to
contain details of the dataset.
-
PREPBIAS
- A bias frame is created using FIGARO MEDSKY to produce the median
of the input images.
-
PREPFLAT
- Uses the bias frame produced by PREPBIAS and a series of input
images to create a flat field using FIGARO MEDSKY. Rotates the images (if
needed) to give horizontal orders.
-
PREPARCS
- Uses the bias frame produced by PREPBIAS and the flat field from
PREPFLAT to prepare the input arc images for ECHOMOP. Images clipped
and rotated.
-
PREPOBJS
- Uses the bias frame produced by PREPBIAS and the flat field from
PREPFLAT to prepare the input object images for ECHOMOP. Images clipped
and rotated.
-
ECHRDARC
- Extracts the arc spectra using an appropriate object image to
determine channels.
-
ECHRDUCE
- Extracts the object spectra.
The PREPBIAS, PREPFLAT, PREPARCS and PREPOBJS scripts use FIGARO commands to
prepare the raw CCD image frames for use by ECHOMOP. By default, these scripts do
not perform rotation of the images. ECHOMOP requires that the orders of the spectrum
should run roughly parallel to the rows of the image (i.e. horizontally). If your input frames
contain échelle spectra with the dispersion running roughly along the columns you should
uncomment the parts of the scripts which deal with image rotation. See the individual scripts for
details.
As FIGARO is used for the data preparation, there is no automatic handling of error values. The
scripts might be adapted to use CCDPACK to perform the CCD-related preprocessing if variances are
required, e.g. for optimal extractions.
The script PREPRUN1 is provided as an example of a driver script for the other tasks. It may be more
convenient to use a non-interactive driver script which you should edit to contain the details of the
dataset. PREPRUN2 is a template for this purpose.
In use, at least a single object and arc should be reduced manually using echmenu to ensure that correct
general parameters are used to build an ECHOMOP reduction file which can be re-used in subsequent
reductions. The scripts ECHRDARC and ECHRDUCE should be edited to reflect the sequence of
operations required for your particular data.
Which parts of a reduction can be fully automated, which parts need to be done only once in the
template manual reduction, which parts must be done manually for every dataset, will depend
entirely on your data. If you are in any doubt, you should study the Introduction to Echelle
Spectroscopy to get an idea of which parts of the reduction are tricky. In theory, ECHOMOP
can be used to perform fully-automated reductions in most cases. In practice, there is no
substitute for carefully investigating your data before trying to ‘pipeline’ process it. Even if you
are confident of successful automated reduction, you will still have to review the results
carefully.
In the scripts included, the echmenu option sequence:
is executed. See SUN/152 for details of ECHOMOP reductions. The EXIT option must be included
otherwise echmenu will revert to interactive mode when the sequence of options is completed. The
reduction sequence is specified via the parameter tune_automate. Other parameters, e.g.
which extraction object to output (parameter result_type), are also given in the echmenu
command line. As the above sequence does not perform order tracing, the order trace
polynomials must be provided in the manually-prepared reduction structure file. This is
an example of an automated reduction sequence where one aspect of the reduction – the
order tracing – has been done only once manually and reused in the subsequent automatic
reductions.
3.5.1 PREPRUN1 & PREPRUN2 – Set up automated run
These are example master driver scripts for an automated run.
If you want to perform optimal extractions or need variance data with from reduction, you’ll need to
know the noise and gain details of the detector used.
Some sort of image display and interrogation programs (e.g. FIGARO IMAGE and ICUR) to work out
which part of the overscan to use for ‘zero-offset correction’ and which part of the frames you want to
keep for extracting the spectra.
Two examples are provided, PREPRUN1 prompts for all the parameters required – in practice you’ll
be better off using PREPRUN2 as there are rather a lot of parameters. The best way to manage
reductions is to take a copy of PREPRUN2 and enter in the script the various trimming numbers, CCD
gain and output noise, and object & arc lists and so on. The whole reduction can then be done by
running the script without any prompts.
Here is the prologue for PREPRUN2:
-
Purpose:
-
Driver script to set up for an automated échelle data reduction run.
-
Language:
-
C shell script.
-
Description:
-
This script can be used to coordinate the relatively complex series of operations required
for reducing a large number of similar échelle spectrograms. Before using the script, you
should be familiar with the use and parameters of the ECHOMOP package. (You can try
this out without being familiar with ECHOMOP – but it is a complex package!)
Essentially the procedure is to ‘prototype’ the reduction manually using ECHOMOP
and then, once you have determined suitable parameter settings, to use the
manually-generated reduction structure file as a template with which to reduce the
complete dataset.
Which parts of the reduction procedure need to be done for every frame, and which
parts can be copied ‘as-is’ from the manual template will depend on your data. For
example, you might use the order traces from the manual reduction for all the frames.
This will be fine as long as the image of the echellogram remains stable over the full time
period which covers your dataset. One way to check this sort of thing is to perform two
manual reductions – one from early in the time period covered, one from late – and then
compare the two. Plotting orders from the reduced arcs is a good way to spot shifts in
the dispersion direction. Detecting shifts in the spatial direction can be more difficult;
however, you might use the tracepoly script to extract the parameters of the order traces
from the ECHOMOP reduction structure files. You can then compare the parameters and
look for shifts – for POLY fits checking that the low-order parameters closely match and
that higher-order parameters are small should be enough.
This script calls a set of shortish scripts to perform each of the data preparation tasks –
debiasing, flat fielding, clipping and so on. You should review the descriptions in these
scripts so that you are happy you understand what each one is doing with your data. You
may also need to edit the scripts in some places, particularly if your echellograms have
orders which run roughly vertically.
The output of the automated reduction process is a series of files ob_‘file’ where ‘file’ is
the name of the source object frame. Arc frames ar_‘file’ are similarly created.
-
Usage:
-
You need to know the true detection area of the CCD used to acquire your data – if you
don’t, display an image with FIGARO IMAGE and look for empty parts of the frame
at the edges of the image. These are not used by ECHOMOP and should be cut off by
setting suitable values for the trim parameters. You should select a part of the overscan
(‘dark’ area) to be used for measurement of the electronic bias level. Use FIGARO ICUR
to measure the coordinates of the various areas.
The following in this script should be edited to suit your data:
-
(1)
- Detector overscan sample region – for bias-level determination.
-
(2)
- Detector clipping region (to remove overscan) – to remove non-science data areas of
the input images.
-
(3)
- Detector output details (noise, gain).
-
(4)
- List of bias frames.
-
(5)
- List of flat-field frames.
-
(6)
- List of arc frames.
-
(7)
- List of arc mask frames (paired with arcs) – these are used to configure the processing
of arcs so that they are extracted in the same way as objects.
-
(8)
- List of object frames.
-
(9)
- Name of prototype ECHOMOP reduction structure file.
See comments in the script for details. Example values have been given for some of these
items.
-
Source code:
-
In a standard Starlink installation the source code for the PREPRUN scripts can be found in
these files:
/star/examples/sc3/preprun1.
/star/examples/sc3/preprun2.
3.5.2 PREPBIAS – Prepare bias frame
-
Purpose:
-
Script to prepare a single biasframe from a series of frames.
-
Language:
-
C shell script.
-
Description:
-
This script produces a single median image from a series of ‘raw’ CCD bias frames.
The median bias frame is created using FIGARO MEDSKY. The output image is called
biasframe, this can be altered by editing the appropriate line in the script.
-
Usage:
-
This script can simply be invoked from the shell; in this case the script will prompt for a
list of the input bias images. Alternatively, the list of bias frames can be supplied on the
command line, for example:
% prepbias run0800 run0801 run0802 run0856 run0857 run0858
If wildcarding facilities are available in your shell, you can use them to simplify the command
line, for example, the above would become:
% prepbias run080[012] run085[678]
in the C shell. This wildcarding facility is available when the script prompts for a list of input
images.
-
Source code:
-
In a standard Starlink installation the source code for PREPBIAS can be found in the
file:
/star/examples/sc3/prepbias
-
Notes:
-
-
(1)
- If needed, the input parameters can be input at the command line thus:
% nohup prepbias filename [filename...] &
the nohup command will ensure that the script continues to run even when you
have logged off the system. The & at the end of the line will run the script in the
background.
-
(2)
- This script is designed to be used as part of an automated échelle data reduction package.
If you intend to use it for this purpose, you should not change the name of the output
median bias frame biasframe. See the comments in the script for changes which can be
made if it is to be used stand-alone.
-
(3)
- This script will work with FIGARO v5.0-0 or later.
-
(4)
- This script is designed to be called by a master reduction script. See the example scripts
preprun1 and preprun2 for details.
3.5.3 PREPFLAT – Prepare Flat-field frame
-
Purpose:
-
Script to prepare a median flat-field for use by ECHOMOP.
-
Language:
-
C shell script.
-
Description:
-
This script takes a list of ‘raw’ CCD flat-field frames and produces a single median
flat-field image suitable for use as the FFIELD file in ECHOMOP.
Be advised that flat-fielding in échelle data reduction is not easy – sometimes it’s not
even possible. Refer to the Introduction to Echelle Spectroscopy (SG/9) if in any doubt.
The script performs the basic CCD data processing steps of bias subtraction and
trimming. Bias subtraction removes the zero-point bias introduced by the camera
electronics. Trimming removes the pre-scan and over-scan regions of the CCD image
which contain no science data and can confuse the algorithms in échelle data reduction
software.
If required, the CCD frames can be rotated so that the dispersion direction of the échelle
orders runs horizontally as required by ECHOMOP.
The median flat-field is calculated using FIGARO MEDSKY.
-
Usage:
-
This script can simply be invoked from the shell; in this case the script will prompt for a
list of the input flat-field images. Alternatively, the list of frames can be supplied on the
command line, for example:
% prepflat run0800 run0801 run0802 run0856 run0857 run0858
If wildcarding facilities are available in your shell, you can use them to simplify the command
line, for example, the above would become:
% prepflat run080[012] run085[678]
in the C shell. This wildcarding facility is available when the script prompts for a list of input
images.
In practice, invocation from your shell is unlikely to be a good method of using this script as 8
environment variables defining the region of the image to be kept and the region of the
overscan to be used for debiasing are required. Use of these variables is summarised
below.
-
Source code:
-
In a standard Starlink installation the source code for PREPFLAT can be found in the
file:
/star/examples/sc3/prepflat
-
Notes:
-
-
(1)
- If needed, the input parameters can be input at the command line thus:
% nohup prepflat filename [filename...] &
the nohup command will ensure that the script continues to run even when you
have logged off the system. The & at the end of the line will run the script in the
background.
-
(2)
- This script is designed to be used as part of an automated échelle data reduction package.
If you intend to use it for this purpose, you should not change the name of the output
median flat-field frame flatfield. See the comments in the script for changes which can be
made if it is to be used stand-alone.
-
(3)
- This script will work with FIGARO v5.0-0 or later.
-
(4)
- When this script is invoked, 8 environment variables defining the overscan region to be
used for debiasing, and the region of the images containing science data must be defined.
These environment variables are used:
-
$xbimin
- X-start of overscan region to use for bias subtract.
-
$xbimax
- X-end of overscan region to use for bias subtract.
-
$ybimin
- Y-start of overscan region to use for bias subtract.
-
$ybimax
- Y-end of overscan region to use for bias subtract.
-
$xtrmin
- X-start of region of image to be retained.
-
$xtrmax
- X-end of region of image to be retained.
-
$ytrmin
- Y-start of region of image to be retained.
-
$ytrmax
- Y-end of region of image to be retained.
-
(5)
- A file biasframe containing a bias frame prepared by the script
prepbias should exist in
the working directory. You can alter the name of this file, see comments in the
script.
-
(6)
- The input frames are not rotated by this script. You may have images in which the orders
run roughly vertically, in which case you should uncomment the line using FIGARO
IROT90 as in the script. Approximately horizontal orders are required by ECHOMOP. If
you do rotate the flat field, note that the script only rotates the final median frame, not the
individual input frames (saves time).
-
(7)
- This script is designed to be called by a master reduction script. See the example scripts
preprun1 and preprun2 for details.
3.5.4 PREPARCS – Prepare arc frames
-
Purpose:
-
Script to prepare a set of arc frames for use by ECHOMOP.
-
Language:
-
C shell script.
-
Description:
-
This script takes a list of ‘raw’ CCD arc-lamp frames and performs the basic CCD
data processing steps of bias subtraction and trimming on the images. Bias subtraction
removes the zero-point bias introduced by the camera electronics. Trimming removes the
pre-scan and over-scan regions of the CCD image which contain no science data and can
confuse the algorithms in échelle data reduction software.
If required, the CCD frames can be rotated so that the dispersion direction of the échelle
orders runs horizontally as required by ECHOMOP.
The script produces a series of files a_‘file’ where ‘file’ is the name of the source frame.
-
Usage:
-
This script can simply be invoked from the shell; in this case the script will prompt for a
list of the input arc-lamp images. Alternatively, the list of frames can be supplied on the
command line, for example:
% preparcs run0800 run0801 run0802 run0856 run0857 run0858
If wildcarding facilities are available in your shell, you can use them to simplify the command
line, for example, the above would become:
% preparcs run080[012] run085[678]
in the C shell. This wildcarding facility is available when the script prompts for a list of input
images.
In practice, invocation from your shell is unlikely to be a good method of using this script as 8
environment variables defining the region of the image to be kept and the region of the
overscan to be used for debiasing are required. Use of these variables is summarised
below.
-
Source code:
-
In a standard Starlink installation the source code for PREPARCS can be found in the
file:
/star/examples/sc3/preparcs
-
Notes:
-
-
(1)
- If needed, the input parameters can be input at the command line thus:
% nohup preparcs filename [filename...] &
the nohup command will ensure that the script continues to run even when you
have logged off the system. The & at the end of the line will run the script in the
background.
-
(2)
- This script is designed to be used as part of an automated échelle data reduction package.
If you intend to use it for this purpose, you should not change the name of the output
frames from a_‘file’ where ‘file’ is an input frame. See the comments in the script for
changes which can be made if it is to be used stand-alone.
-
(3)
- This script will work with FIGARO v5.0-0 or later.
-
(4)
- When this script is invoked, 8 environment variables defining the overscan region to be
used for debiasing, and the region of the images containing science data must be defined.
These environment variables are used:
-
$xbimin
- X-start of overscan region to use for bias subtract.
-
$xbimax
- X-end of overscan region to use for bias subtract.
-
$ybimin
- Y-start of overscan region to use for bias subtract.
-
$ybimax
- Y-end of overscan region to use for bias subtract.
-
$xtrmin
- X-start of region of image to be retained.
-
$xtrmax
- X-end of region of image to be retained.
-
$ytrmin
- Y-start of region of image to be retained.
-
$ytrmax
- Y-end of region of image to be retained.
-
(5)
- A file biasframe containing a bias frame prepared by the script
prepbias should exist in
the working directory. You can alter the name of this file, see comments in the
script.
-
(6)
- The input frames are not rotated by this script. You may have images in which the orders
run roughly vertically, in which case you should uncomment the line using FIGARO
IROT90 as indicated in the script. Approximately horizontal orders are required by
ECHOMOP.
-
(7)
- This script is designed to be called by a master reduction script. See the example scripts
preprun1 and preprun2 for details.
3.5.5 PREPOBJS – Prepare object frames
-
Purpose:
-
Script to prepare a set of object frames for use by ECHOMOP.
-
Language:
-
C shell script.
-
Description:
-
This script takes a list of ‘raw’ CCD object frames and performs the basic CCD data
processing steps of bias subtraction and trimming on the images. Bias subtraction
removes the zero-point bias introduced by the camera electronics. Trimming removes the
pre-scan and over-scan regions of the CCD image which contain no science data and can
confuse the algorithms in échelle data reduction software.
If required, the CCD frames can be rotated so that the dispersion direction of the échelle
orders runs horizontally as required by ECHOMOP.
The script produces a series of files o_file where file is the name of the source frame.
-
Usage:
-
This script can simply be invoked from the shell; in this case the script will prompt for
a list of the input object images. Alternatively, the list of frames can be supplied on the
command line, for example:
% prepobjs run0800 run0801 run0802 run0856 run0857 run0858
If wildcarding facilities are available in your shell, you can use them to simplify the command
line, for example, the above would become:
% prepobjs run080[012] run085[678]
in the C shell. This wildcarding facility is available when the script prompts for a list of input
images.
In practice, invocation from your shell is unlikely to be a good method of using this script as 8
environment variables defining the region of the image to be kept and the region of the
overscan to be used for debiasing are required. Use of these variables is summarised
below.
-
Source code:
-
In a standard Starlink installation the source code for PREPOBJS can be found in the
file:
/star/examples/sc3/prepobjs
-
Notes:
-
-
(1)
- If needed, the input parameters can be input at the command line thus:
% nohup prepobjs filename [filename...] &
the nohup command will ensure that the script continues to run even when you
have logged off the system. The & at the end of the line will run the script in the
background.
-
(2)
- This script is designed to be used as part of an automated échelle data reduction package.
If you intend to use it for this purpose, you should not change the name of the output
frames from o_‘file’ where ‘file’ is an input frame. See the comments in the script for
changes which can be made if it is to be used stand-alone.
-
(3)
- This script will work with FIGARO v5.0-0 or later.
-
(4)
- When this script is invoked, 8 environment variables defining the overscan region to be
used for debiasing, and the region of the images containing science data must be defined.
These environment variables are used:
-
$xbimin
- X-start of overscan region to use for bias subtract.
-
$xbimax
- X-end of overscan region to use for bias subtract.
-
$ybimin
- Y-start of overscan region to use for bias subtract.
-
$ybimax
- Y-end of overscan region to use for bias subtract.
-
$xtrmin
- X-start of region of image to be retained.
-
$xtrmax
- X-end of region of image to be retained.
-
$ytrmin
- Y-start of region of image to be retained.
-
$ytrmax
- Y-end of region of image to be retained.
-
(5)
- A file biasframe containing a bias frame prepared by the script
prepbias should exist in
the working directory. You can alter the name of this file, see comments in the
script.
-
(6)
- The input frames are not rotated by this script. You may have images in which the orders
run roughly vertically, in which case you should uncomment the line using FIGARO
IROT90 as indicated in the script. Approximately horizontal orders are required by
ECHOMOP.
-
(7)
- This script is designed to be called by a master reduction script. See the example scripts
preprun1 and preprun2 for details.
3.5.6 ECHRDARC – Reduce arc frames
-
Purpose:
-
Script to reduce an arc frame with ECHOMOP.
-
Language:
-
C shell script.
-
Description:
-
This script reduces a wavelength-scale reference arc frame using ECHOMOP. An object
or flat-field frame to be used for order profiling must be available. (You could use an arc
frame – this is fine as long as the same frame is used by the script echrduce for reducing
the object frames for which this is the arc reference.)
The output file is named ar_ArcFile where ArcFile is the name of the input arc frame.
-
Usage:
-
This script can simply be invoked from the shell; in this case the script will prompt
for the arc frame to be processed and the object frame to be used for order profiling.
Alternatively, the input frame names can be supplied on the command line, for example:
In practice, invocation from your shell is unlikely to be a good method of using this script as
3 environment variables defining the CCD output characteristic and ECHOMOP
reduction structure file name are required. Use of these variables is summarised
below.
-
Arguments:
-
If supplied, command-line arguments must be in this order:
-
(1)
- Arc Frame. Name of the Arc frame to be processed.
-
(2)
- Object Frame. Name of the Object frame to be processed.
Missing command-line arguments are prompted for.
-
Source code:
-
In a standard Starlink installation the source code for ECHRDARC can be found in the
file:
/star/examples/sc3/echrdarc
-
Notes:
-
-
(1)
- If needed, the input parameters can be input at the command line thus:
% nohup echrdarc arcfilename objectfilename &
the nohup command will ensure that the script continues to run even when you
have logged off the system. The & at the end of the line will run the script in the
background.
-
(2)
- This script is designed to be used as part of an automated échelle data reduction package.
If you intend to use it for this purpose, you should not change the name of the output arc
frame, ar_‘ArcFile’.
-
(3)
- When this script is invoked, 3 environment variables defining the output characteristics of
the CCD used, and the ECHOMOP reduction structure file used must be defined. These
environment variables are used:
-
$EchFile
- Name of the ECHOMOP reduction structure file.
-
$Gain
- CCD output transfer function in photons per ADU.
-
$RDN
- CCD readout noise in electrons.
-
(4)
- A file flatfield containing a flat-field frame prepared by the script
prepflat should exist
in the working directory. You can alter the name of this file, see comments in the
script.
-
(5)
- The scripts
preparcs, prepbias and prepflat should be used to prepare data for
processing with this script.
-
(6)
- This script is designed to be called by a master reduction script. See the example scripts
preprun1 and preprun2 for details.
3.5.7 ECHRDUCE – Reduce object frames
-
Purpose:
-
Script to reduce a set of object frames with ECHOMOP.
-
Language:
-
C shell script.
-
Description:
-
This script reduces a series of object frames using ECHOMOP.
The output files are named ob_File where File is the name of the input object frame.
-
Usage:
-
This script can simply be invoked from the shell; in this case the script will prompt for a
list of object frames to be processed Alternatively, the input frame names can be supplied
on the command line, for example:
% echrduce run0800 run0801 run0802 run0856 run0857 run0858
If wildcarding facilities are available in your shell, you can use them to simplify the command
line, for example, the above would become:
% echrduce run080[012] run085[678]
in the C shell. This wildcarding facility is available when the script prompts for a list of input
images.
In practice, invocation from your shell is unlikely to be a good method of using this script as
3 environment variables defining the CCD output characteristic and ECHOMOP
reduction structure file name are required. Use of these variables is summarised
below.
-
Arguments:
-
If supplied, command-line arguments must be in this order:
-
(1)
- List of files. List of files to be processed.
Missing command-line arguments are prompted for.
-
Source code:
-
In a standard Starlink installation the source code for ECHRDUCE can be found in the
file:
/star/examples/sc3/echrduce
-
Notes:
-
-
(1)
- If needed, the input parameters can be input at the command line thus:
% nohup echrduce filename [filename...] &
the nohup command will ensure that the script continues to run even when you
have logged off the system. The & at the end of the line will run the script in the
background.
-
(2)
- This script is designed to be used as part of an automated échelle data reduction package.
If you intend to use it for this purpose, you should not change the name of the output
frames, ob_‘File’.
-
(3)
- When this script is invoked, 3 environment variables defining the output characteristics of
the CCD used, and the ECHOMOP reduction structure file used must be defined. These
environment variables are used:
-
$EchFile
- Name of the ECHOMOP reduction structure file.
-
$Gain
- CCD output transfer function in photons per ADU.
-
$RDN
- CCD readout noise in electrons.
-
(4)
- A file flatfield containing a flat-field frame prepared by the script
prepflat should exist
in the working directory. You can alter the name of this file, see comments in the
script.
-
(5)
- The scripts
prepobjs, prepbias and prepflat should be used to prepare data for
processing with this script.
-
(6)
- This script is designed to be called by a master reduction script. See the example scripts
preprun1 and preprun2 for details.
Copyright © 2000, 2003 Council for the Central Laboratory of the Research Councils