In this section, a simple run-through of the extraction process using Starlink software is given. §8 gives answers to specific questions and solutions to common problems.
Before you start the extraction you will have to do the detector-specific preparation of your data (most likely to remove CCD-related effects). If you have not done this, refer to §4.1 for an outline of the procedure, and §1.2 for pointers to documentation of the process.
The first thing you need to do (if you’ve managed to prepare your CCD data you will most likely know this already, but…) is to run the Starlink setup. Normally Starlink software is installed in the /star directory and the commands you must execute are:
If your Starlink software is installed somewhere other than /star—one place to look is STARLINK_DIR—then modify the commands appropriately. Most people include these lines in their shell login file (.login in your home directory).
Once you have done the Starlink ‘login’ you can initialise for any of the major packages simply by typing their names. For example, we are going to use kappa[8] and echomop[16] and so get a display something like:
We can now use kappa commands and echmenu, the menu-driven front-end for echomop.
At this point you might like to take a copy of the example data which comes with this document when installed as part of the Starlink document set. You will find the files in the directory
Create an empty directory and enter it using cd. To copy the test data type the command
You will then find you have these datafiles in your directory:
| File | Description |
object.sdf | Frame with the object spectrum |
flat.sdf | Frame with a flat field |
arc.sdf | Frame with a wavelength-reference arc |
object1.sdf | Frame with a rotated spectrum |
The first thing you need to do is ensure that the images are oriented correctly. To do this use kappa display to look at the image:
This will display the image object using a false-colour look-up table. You may have to use the xdisplay[2] command to ensure that the image is sent to the correct (i.e. your) screen. display has many options, for the moment we just need to check the orientation of the image, the accept keyword tells kappa to use default values for the parameters we have not given. The example data are correctly oriented, with the spectrum dispersion from left-to-right.
Try
The clear parameter indicates that we want the previous display to be erased before object1 appears. You’ll see that this spectrum looks similar to the previous one, but it runs from top-to-bottom of the image—it needs to be rotated. To do this, use the kappa command rotate.
This rotates the data in object1 clockwise by 90 degrees and places the result in object1r. If you find that your real data need to be rotated, you will have to apply the change to all the data files (including arcs and flat fields). Try this to inspect the rotated data:
Jumping ahead (quite a way) you may want to ensure that the now correctly rotated data are arranged so that the spectrum is dispersed with wavelength increasing from left-to-right. Don’t worry if you don’t know how to do this—it isn’t strictly necessary; however, you may find it easier to recognise the arc spectrum in an atlas if it is arranged with the wavelength increasing left-to-right as this is the way the atlases are printed. You can always come back later and flip the images if you find the arcs hard to recognise. There’s no single way to spot which way the spectra are dispersed. You have to know a little about the spectra you are working with; where the strong or large features are. You might look at the arc as well for some familiar pattern in the reference spectrum.
If you do find that you need to reverse the dispersion direction, you can use kappa flip to do this:
This tells kappa to flip the first (X-) axis in the data and put the resulting image in the dataset object1rf. You can see here the common practice of appending letters to the dataset names to indicate what processing they have undergone—‘r’ for rotate, ‘f’ for flip. This can be helpful if you have many images and are going to work over several sessions.
We can now proceed to the first stage in the actual spectrum extraction.
The spectrum extraction is done using the echomop package. At this stage, you will find it easiest to work through the example extraction here, rather than trying to understand all the facilities and parameters of echomop.
The first stage is to start echmenu:
Each reduction is described by a database file specified by the ech_rdctn parameter, in this case we
have chosen rdf1, which the program creates. The display parameter tells echmenu to overlay trace
plots on the image itself so we can see how well the trace is doing. The soft parameter points output
graphs to our existing xwindows screen. Don’t worry about tune_mxskypix for now, it is needed (and
explained) later on.
echmenu displays its complete menu—most of the options are specialised and we don’t need them at this stage. echmenu works through the extraction process in a series of numbered steps (1, 2, 3, etc…). You can exit echmenu between stages and return later to pick up the reduction at the same point. echmenu tends to be quite verbose when displaying messages—don’t be put off by this, most of the information is not useful at this stage and can simply be ignored. For brevity, most of these messages are not included in future screen display listings.
The prompt at the end of the above display is suggesting Option 1 Start a reduction which seems like a good place to begin:
There are three prompts which appear: first, tracim is asking which image to use to trace the path of the spectrum—in this case we are able to use the object itself; inptim is asking for the name of the file from which the spectrum will be extracted, again this is object; lastly, arc is the name of the wavelength-reference image, which is called arc in the example data.

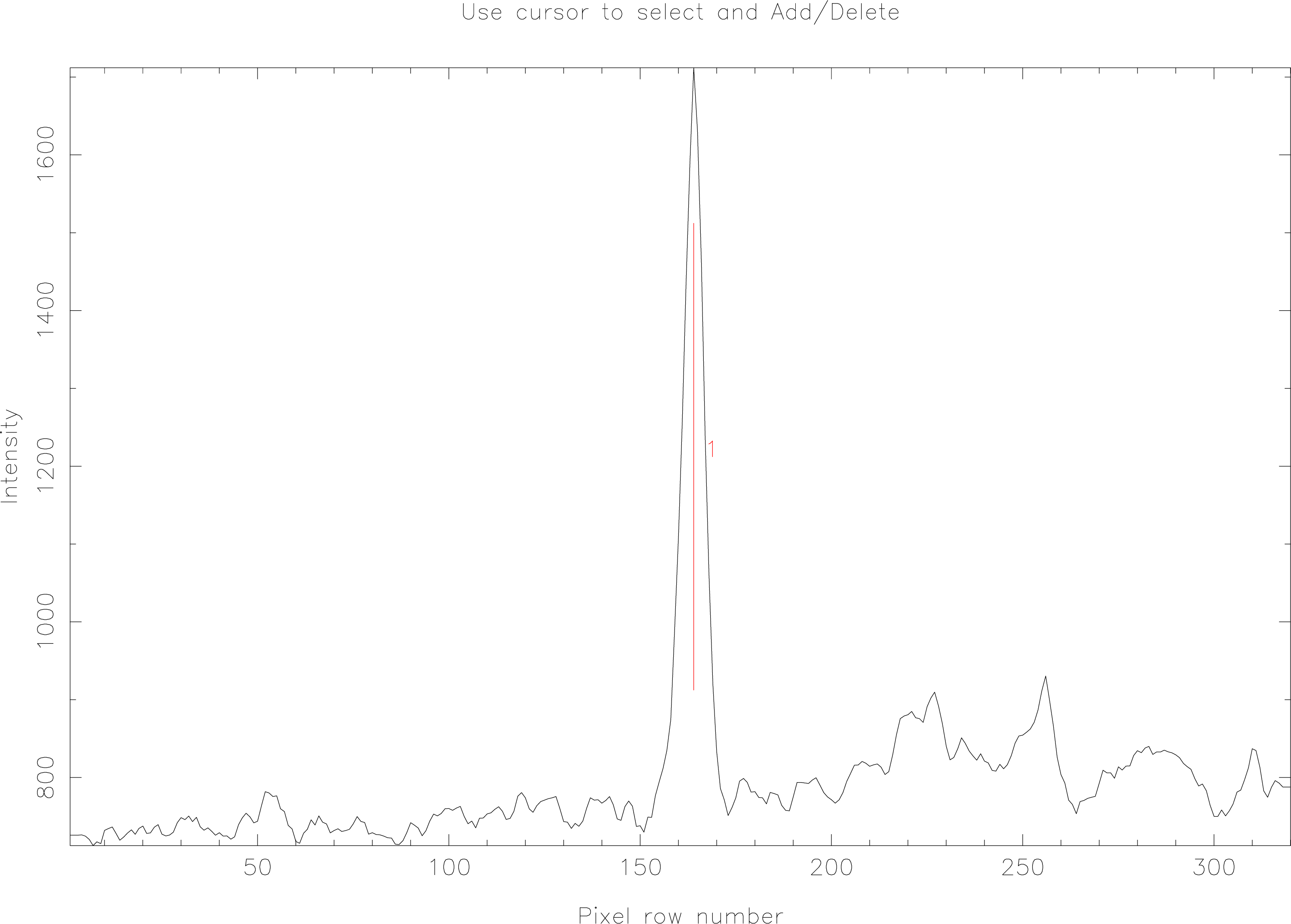
echmenu now displays a slice across the image object and you will see that it automatically locates features which look like they may be spectra. Figure 10 shows such a plot.
A short menu of options is displayed, if (as in this case) all is well, you can just hit the E key to accept the spectrum as correctly found. You may find that echmenu incorrectly guesses where the spectrum is; you can use the C key to remove the automatically located spectra and the A key to add a spectrum at the point indicated by the graphics cursor.
You will notice that echmenu appears to be able to handle more than one spectrum in one image; this is the case—it can handle multiple separate spectra (e.g. from a fibre spectrograph) and multi-order spectra from échelle spectrographs. But we don’t need to know about that at the moment.
Having located the spectrum, we now get another menu; this is in fact a shortened version of the full menu with the ‘most-likely’ choices at the top of the list. We want the second option Trace orders; we only have one order but it works the same way.
The parameters here are: trcfit is the type of fit function to use, poly is often fine, spline works in
most other cases; trace_mode determines which algorithm is used to find the spectrum at each
X-sample (i.e. along the wavelength/dispersion axis), C means centroid and usually works fine, other
options include G for a Gaussian fit, and B for a simple centre-of-gravity fit; trc_npoly selects the
number of fit parameters (i.e. the order of fit for a poly) for a spline you need more parameters (seven
plus number-of-knots, times two).
You will see a line plotted overlaid on the image on the graphics device. (Much like Figure 5, see page 25.) This shows the path of the trace as determined by echmenu. Usually the trace works well; to inspect the fit we use Option 3 Clip fitted traces. The only parameter you will be prompted for here is
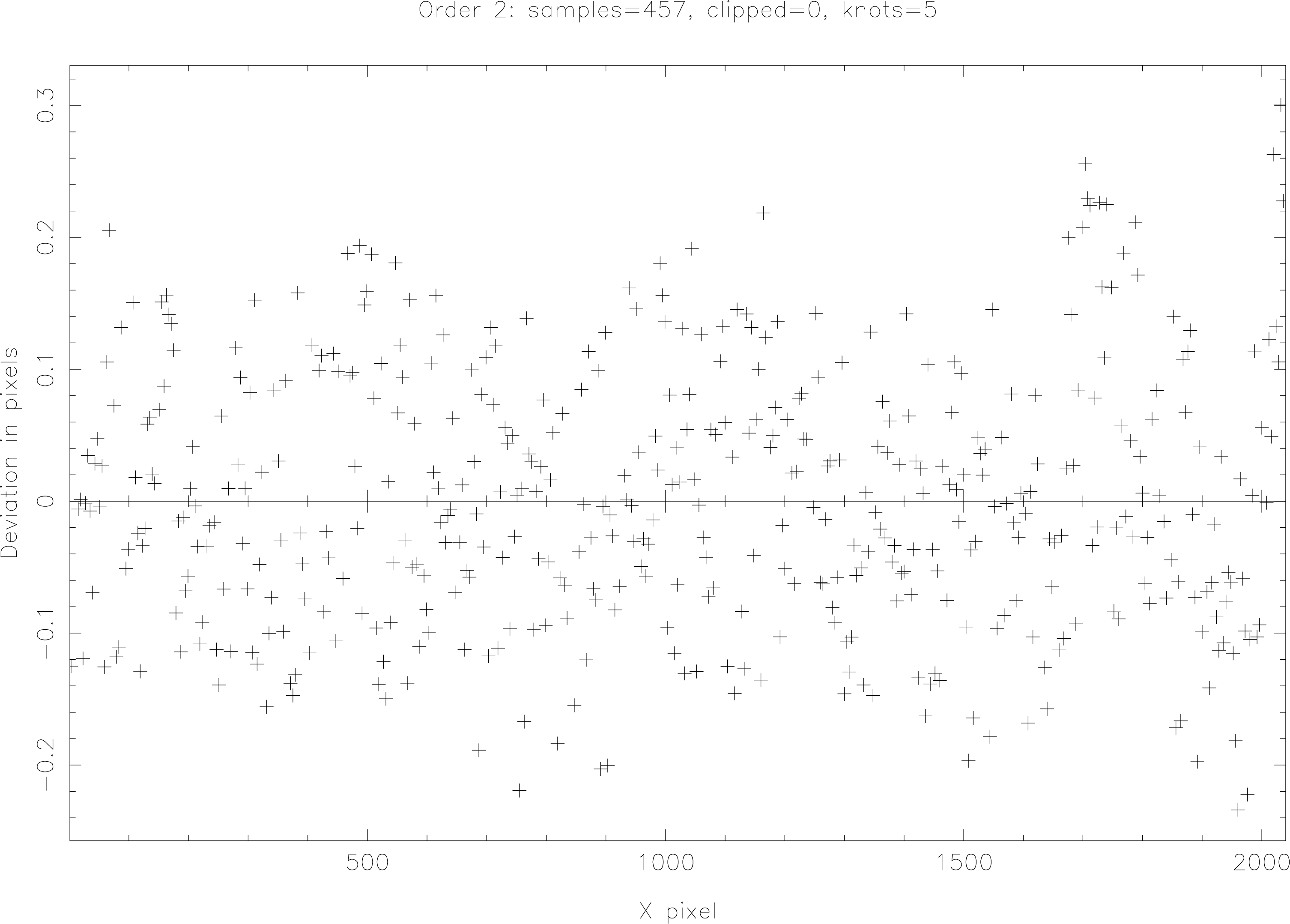
Accept the default value (by hitting return) as we want to be able to improve the trace interactively. The first plot (see Figure 11) shows the deviation of each X-sample in image pixels from the trace path. Values less than half a pixel are fine. To see the trace points plotted on the fitted trace curve hit the V key (the full menu of options in the clipper can be displayed by hitting M). Hit V again to return to the ‘deviations’ plot. Deviant points can be clipped in a number of different ways: use the . key to delete the point closest to the graphics cursor; N clips points below the cursor Y-position; P clips points above the cursor Y-position. Option R is often useful if part of the trace has gone wrong; hit R to mark one end of the bad X-range and then hit the mouse (usually left) button to mark the other end. As points are clipped the display updates and the points disappear—you should see the extent of the deviations reduce.
When clipping, don’t overclip—a nice ‘cloud’ of deviations of up to about plus or minus half of a pixel is fine. If, when using a poly curve you find that a very high-order fit (anything above 7th or, perhaps 8th, order) is needed to get a good fit then you probably should try a spline fit instead as you will start to ‘over-fit’ the data.
The next step in the ‘data-modelling’ process is to take a section through the spectrum (cross dispersion) and decide which pixels contribute to the object signal and which to the background. The process is split into two stages: first, deciding where the edges of the dekker are; second, choosing the channels. Both stages are covered by echmenu Option 4 Determine dekker/object extent, they can be done separately as options 4.1 (dekker setup) and 4.2 (channel setup).
If you didn’t specify tune_mxskypix=31 when you started this echmenu session you should refer to
§8.1 for details of how to do this within echmenu.
The first prompts in Option 4 are:
You should set pfl_interact to yes (or true, means the same thing) as you may want to adjust the
dekker limits interactively. slitim is the name of the image you are going to use to find the
edges of the slit; the object image will do, but the flat-field image is often better as the
flat-field has steeper, and so clearer, edges. Specify flat if you are working with the example
dataset.
As in previous options, you will be presented with a displayed graph and a short menu of options. The graph shows a section across the dispersion generated by averaging the cross-dispersion profile in some fraction of the X-samples. By default echmenu uses the middle 20% of the pixels in the X-direction. This gives a smooth(er) curve than a single-pixel section and avoids the possibility of accidentally choosing a section across a strong absorption feature.

In the plot (see Figure 12) pixels which echmenu has guessed as inside the slit are displayed coloured red2 and in a solid line; pixels outside the slit are in green and a dot-dash line style. To change the upper or lower boundary, position the graphics cursor at the new dekker edge position, and hit either the U key (upper limit) or L key (lower). The new selection will be displayed. If the profile appears to be wider than the displayed graph then click a U or L with the cursor outside the appropriate edge of the outline box and the display will expand. Once you are happy with the selection of the edges of the dekker hit the E key.
The next graph which pops up (such as Figure 4 (see page 22)) is slightly more complex. Once echmenu has the dekker limits it attempts to estimate which pixels within the slit are from the object, and which are background (called sky pixels in echomop). In the plot, pixels chosen as object are shown in red and a solid line; pixels selected as background are in blue and a dashed line; pixels outside the dekker are again shown in green and a dot-dash line. To help indicate the object and background channels, the pixels selected are also marked with horizontal ‘brackets’; the object at the top (red); the background at the bottom (blue).
At this point you should examine the profile carefully, looking for suitable object and background channels. It may be the case that the default echmenu settings automatically select good channels; however, better results are often obtained by manually tweaking the selections. You can still adjust the dekker limits at this point, again using the U and L keys. Use the S key to mark a particular pixel in the profile as background (sky) and O to mark it as object. If you are working with the example datasets, try hitting S with the graphics cursor somewhere near the middle of the profile; you will see the display update and a pixel will change from object to background. Click O to change it back to object.
In the same way as in the trace-clipping option you can use the R key to start marking a range of pixels; press either O or S with the graphics cursor at the other end of the range to mark the pixels as object or background respectively.
You may have some pixels which appear to be neither object nor background—perhaps there is some other object contaminating the slit. Use the I key to mark these pixels to be ignored, such pixels appear in black, solid-line style on the plot.
At any point hitting the M key will display the full menu of options. Hit the E key when the channels look sensible. We now have the basic parts of the model describing the data—a trace, and object and background channels.
echmenu provides many options for flat fielding, these are handled by Option 5 Model flat field.
There are only two prompts which appear at this point. ffield is the name of our flat-field image (in this case flat) you can specify none if you don’t need flat fielding or have no image to use. fltfit is the type of model to fit to the flat field. We have selected median as this gives better rejection of deviant data than mean, which simply uses the average value over nearby pixels to generate a smooth fit to the continuum lamp spectrum. There are many other options, and the fit can be under interactive control. A simple model is usually fine.
The next stage in spectrum extraction is to model the background. echmenu Option 6 Model sky
performs the modelling and you can accept the default values for the parameter prompts
and get a reasonable model. If you intend to perform optimal extraction you will need to
find the photon_to_adu (gain) value and readout_noise. You can do this using kappa
fitslist to list the FITS extension in the input dataset, e.g., for the object in the example
data:
You should find the appropriate CCD parameters in the header, see §8.4 for more details. If the information you need is not present in the FITS headers you will have to consult the observatory’s CCD manual.
The only other data required prior to extraction is the profile model, and this is only needed for optimal extractions. echmenu Option 7 Model object profile will generate a suitable model. If you have been working through the example reduction data in a single session, echmenu will not prompt you for any further parameters when you run Option 7.
And so to Option 8, Extract orders 1-D. The only new parameter for this option is
The default value, O, means optimal extraction. Other options are: S for a simple unweighted extraction, and P for a profile-weighted extraction.
echmenu has an Option (number 27) for inspecting the contents of the current reduction database file. You can start the option by typing P (for ‘plot’) at the main-menu prompt. Once in the plotter, a menu appears and new prompt:
If you type obj (for object) and then hit return, you will get a plot of the extracted object spectrum (see Figure 7, page 31, for an example). You can get a list of things you can plot by typing D (for directory). Here’s some of the ones which might interest you at this stage:
| You type | You see | You type | You see |
SKY | Sky spectrum | SKYV | Sky spectrum errors |
OBJ | Extracted object spectrum | OBJV | Extracted object errors |
ARC | Extracted arc spectrum | FWAV | Fitted wavelengths |
FFLT | Fitted flat-field balance factors | ||
We haven’t generated the wavelength scale, so fwav isn’t yet available.
Another useful plotter option is L (limit X- and/or Y-range of plot). You are prompted for the limiting values in both X- and Y-directions (Y as plotted, rather than in the source image—so for OBJ, Y will be flux; for FWAV, Y will be wavelength); enter zero if any limit is to be determined from the data.
Once you have finished inspecting the spectrum, hit Q to return to the main menu.
There are two stages to echmenu wavelength calibration; the first stage is simple; the second stage may be more complex. The first step is to run Option 9 Locate arc line candidates. This searches the arc spectrum for features which look like arc lines. The result is a small list of potential features which can be used in the second stage, wavelength calibration.
echmenu Option 10 Identify features performs wavelength calibration. There are a number of prompts:
Arc lamp database information for CuAr and ThAr lamps is available, if you use some other lamp you
will need a list of wavelengths for features in the spectrum of the lamp. Often a ThAr lamp is used,
however, in the example data the lamp used is CuAr, so you can should set ech_ftrdb=$ARCDIRS/CUAR
and arc_type=$ARCDIRS/CUAR.ARC. Note that the names of these files are case-sensitive. wavfit is the
type of fit to use, poly should be fine.
If you select auto_id=true, echmenu will attempt a fully automatic wavelength calibration. This will
often work; however, it is better to inspect the fit manually, rather than just accepting it automatically;
so we accept the default auto_id=false.
hi_wave and low_wave are limits on the wavelength range that might be covered by the spectrum;
setting the limits both to zero (the default) indicates we have no idea of the wavelength range.
However, if you do have a rough idea of either or both limits then enter them here. Constraining the
wavelength range speeds up the process of feature identification. The units are Angstroms for the
ThAr and CuAr databases.
max_dispersion and min_dispersion are the limits on the dispersion of the spectrum. Again, if you
know the dispersion, set these values close to the value to help constrain automatic feature
identification. Inspection of FITS headers may reveal the dispersion used in your data, you might
otherwise look in the instrument handbook for the spectrograph used. The units here are Angstroms
per detector pixel. The default values are set for common échelle spectrographs and so may be too
small for your data. If in doubt, set max_dispersion to a large value and leave the default for
min_dispersion as it is.
w_npoly is the number of coefficients of fit to use for the wavelength polynomial. The default value of
7 is fine. The fitter will adjust the order automatically the first time a fit is made if the value is
unusable.
Once you have worked through all the prompts for Option 10 another mini-menu will appear offering a default of I (identify features manually). Accept the default by pressing return and (yet) another menu appears. The graphics display will also update showing the arc spectrum. Each potential feature identified in the arc spectrum will be marked by a short vertical dash above the feature. This will be similar-looking to Figure 8 (see page 34) except the features will not yet be labelled with wavelengths.
All these sub-menus may appear confusing, however, you can just accept the default to get to the interactive wavelength calibration. The other options are used when the dataset is multi-spectrum or multi-order. You should now have a menu like this one:
As you can see, there are rather a large number of options. To apply an option, type its first letter with the graphics cursor on the displayed plot. Try an A, which will attempt automatic calibration. You will see some details of the fit displayed and then a plot with the wavelengths of the identified features overlaid. For the example dataset this is all you need to do—you now have a wavelength scale. If the display zooms on to only a small part of the spectrum hit R to get a full-spectrum plot.
At this point you may want to have one of the atlases mentioned earlier to hand to check the identifications manually. You can inspect the line-wavelength lists on-line, for example, the ThAr list by:
The important point in deciding whether the fit is good is the RMS error of the fit. Below is an example of a good fit:
You can see that the RMS error is quite small, also the ‘RMS if omitted values’ are all of similar values. If a mis-identified or badly recorded feature is included, then this will be indicated by a much lower ‘RMS if omitted’ than the other features. To remove such a feature: position the graphics cursor on the feature; hit D to delete the feature from the list of features to be fitted; hit F to apply the new fit.
It is important not to over-fit the feature list. In the above example ten points are fitted with a fourth-order curve; this is fine, seventh-order would be too high as the errors would then be fitted away. The plus and minus (+ and -) keys change the order of fit. Press the E key when you are happy with the fit.
Once you have got the extracted, wavelength-calibrated spectrum you have two options: you can output the data from echomop; or, you can normalise the spectrum to give a more-or-less flat continuum. The next section explains how to output data from echomop. This section outlines using Option 11 Flatten order shape to normalise the data.
Enter 11 at the main echmenu prompt and the following parameter prompts will appear:
ffield is the name of the image to be used to model the instrument profile, if you have been working
through the example data in one session you won’t be prompted for this parameter again. You don’t
have to use a flat-field image; one option is to use the object spectrum image itself and fit a curve to
the continuum. If you are following the example reduction, choose to use the flat-field image
flat. For modelling the instrument response a spline curve is often better than a poly,
this is simply because it is unusual for the response to be well represented by a simple
polynomial, hence we have chosen blzfit=spline. blz_interact=y selects interactive
adjustment of the fit. blz_npoly sets the number of parameters for the fit; we have chosen 28 as
this is a spline. (Remember, for a spline we need seven plus number-of-knots, times two,
parameters.)
After entering values for the parameter prompts, you will see a display very similar to that in the trace clipper; the same bit of code does both jobs. This means that you can use the same keys (e.g. V, to see the fitted curve versus the measured data) to view and improve the fit. Use + and - to increase or decrease the order of fit; C to clip points further from the X-axis than the current graphics cursor position. Once you are happy with the fit, hit E to accept it.
At this point you can re-run Option 27 (or P, the plotter) and view the flattened spectrum:
echmenu Option 14 Save reduced data handles the output of data. You can produce NDFs which dipso[11], kappa and figaro can read, or you can output ASCII listings of the spectra.
Here ech_rducd is the rather opaque-sounding prompt for ‘output file name’; result_format is NDF,
select ASCII for a listing instead; result_type should be set to extobj to output the extracted
spectrum, extarc to output the extracted arc.
If you followed the example data reduction, you will now have a file spectrum.sdf in the working directory which contains a normalised, wavelength-calibrated spectrum which can be read by other Starlink software.
2The colours referred to will only be apparent if your medium/display supports colour.